Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
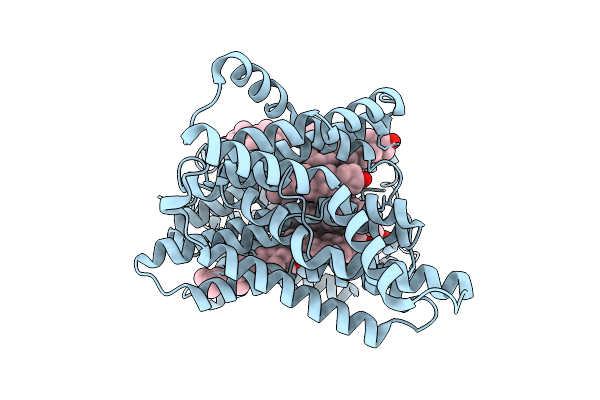 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: CLR |
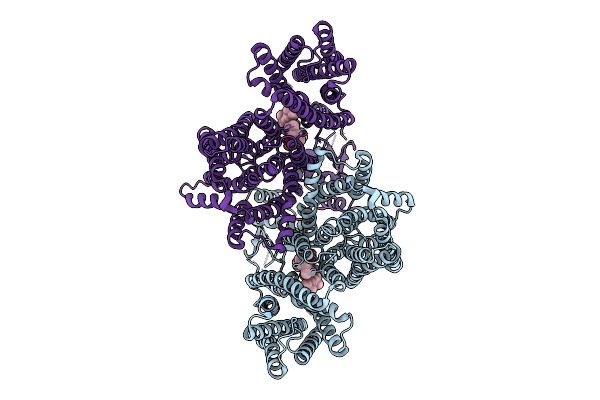 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: Y01, CLR |
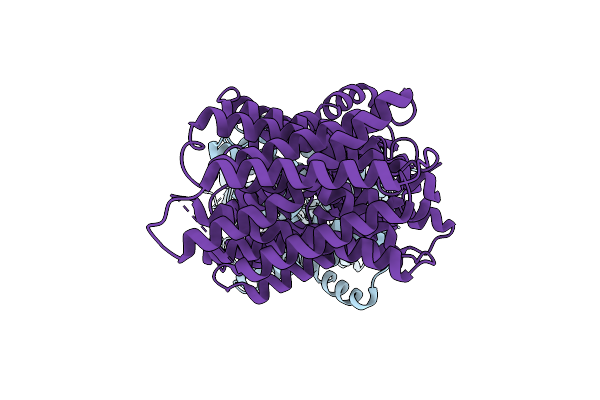 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
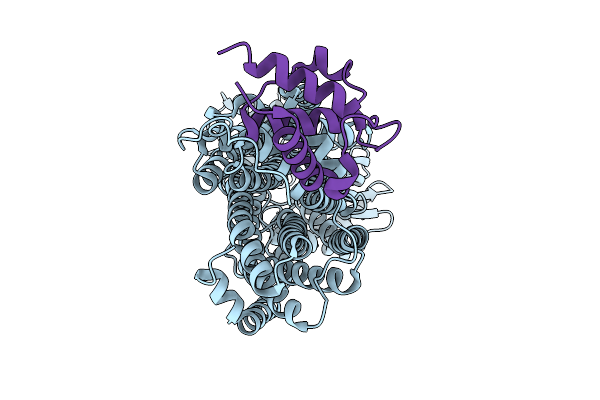 |
Cryo-Em Map Of Msmeg_3496 In Complex With Acpm, Size Exclusion Chromatography Peak1
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
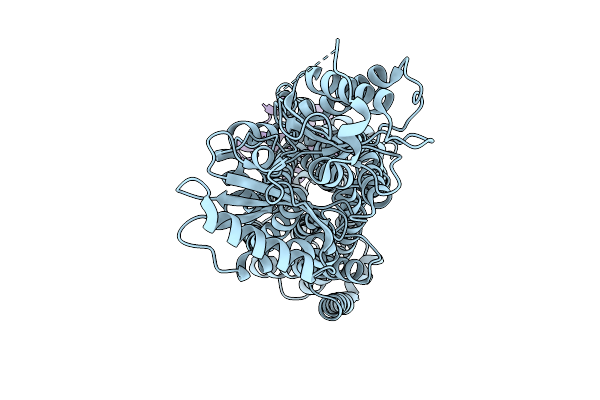 |
Cryo-Em Map Of Msmeg_3496 In Complex With Acpm, Size Exclusion Chromatography Peak2
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
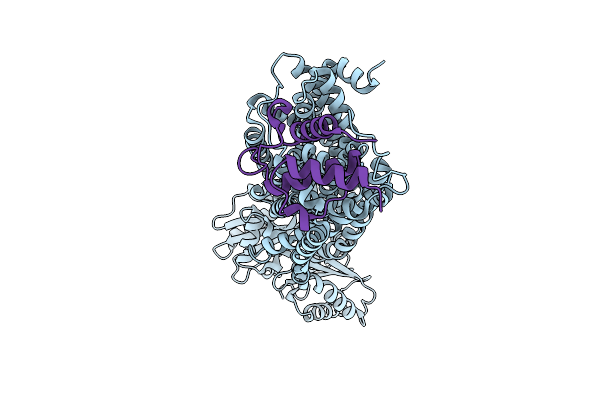 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv), Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
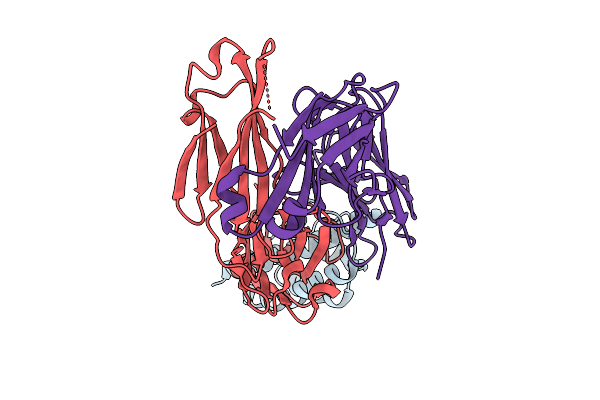 |
Organism: Canis lupus familiaris
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: CYTOKINE/IMMUNE SYSTEM |
 |
Structural Insights Into Il-31 Signaling Inhibition By A Neutralization Antibody
Organism: Canis lupus familiaris
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: CYTOKINE/IMMUNE SYSTEM |
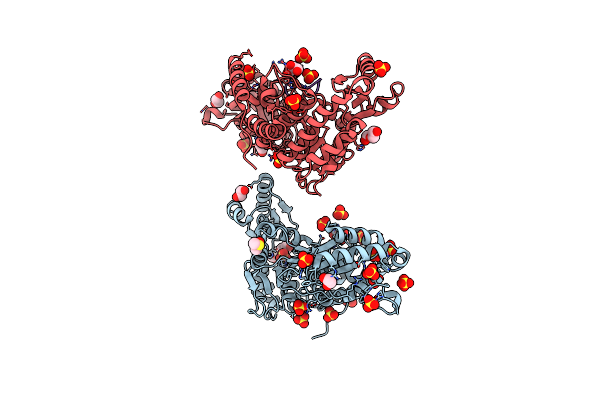 |
Mycobacterium Tuberculosis Pks13 Acyltransferase Serine Converted To Beta-Lactam Form By Cec215 Via Sufex Reaction
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: ANTIBIOTIC Ligands: SO4, CL, PG4, EDO, DMS, GOL, PEG |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: ANTIBIOTIC Ligands: DMS, SO4, PE5, EDO, PEG, GOL, CL, NA, P33 |
 |
M. Tuberculosis Pks13 Acyltransferase (At) Domain In Complex With Sufex Inhibitor Cec215
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 1PE, SO4, A1ATV |
 |
M. Tuberculosis Pks13 Acyltransferase (At) Domain In Complex With Sufex Inhibitor Cmx410
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: TRANSFERASE/TRANSFERSE INHIBITOR Ligands: 1PE, SO4, A1ATW |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: TRANSFERASE Ligands: 1PE, SO4 |
 |
M. Tuberculosis Pks13 Acyltransferase (At) Domain In Complex With Sufex Inhibitor Cmx410 - Reaction Product
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: TRANSFERASE Ligands: SO4, 1PE, A1AVL |
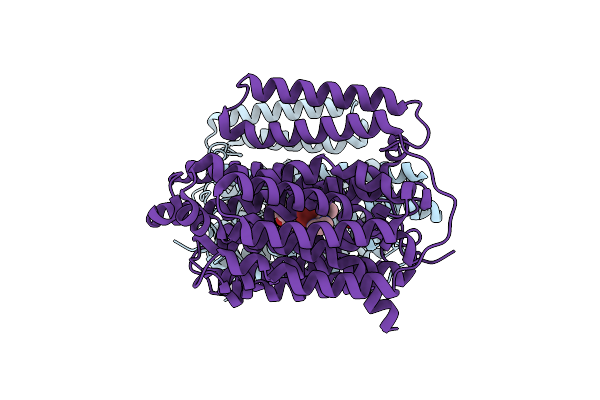 |
Organism: Mycobacterium tuberculosis h37rv
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: PROTEIN BINDING Ligands: A1AQR |
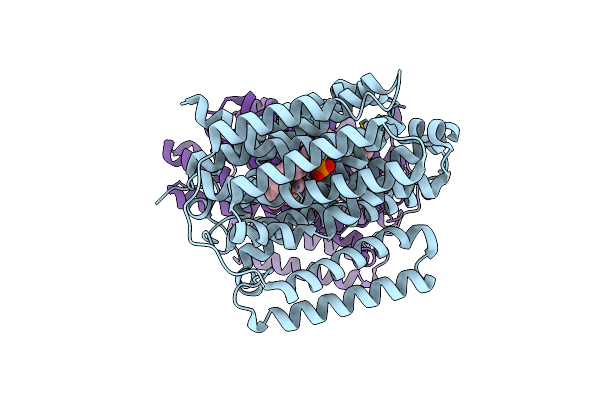 |
Structure Of The Multidrug Efflux Pump Efpa From M. Tuberculosis Complexed With Lipids
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2024-11-27 Classification: MEMBRANE PROTEIN Ligands: Y86, Y8F |
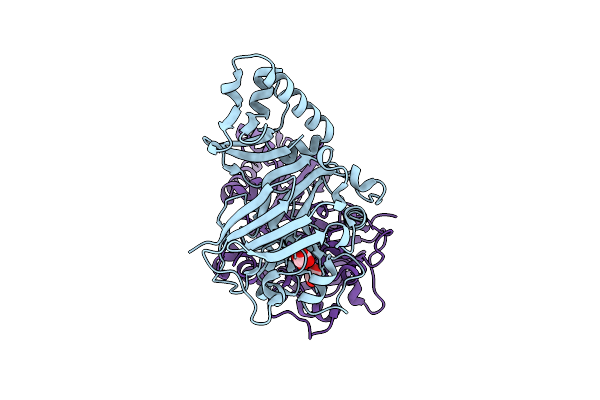 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: OXL, MN |
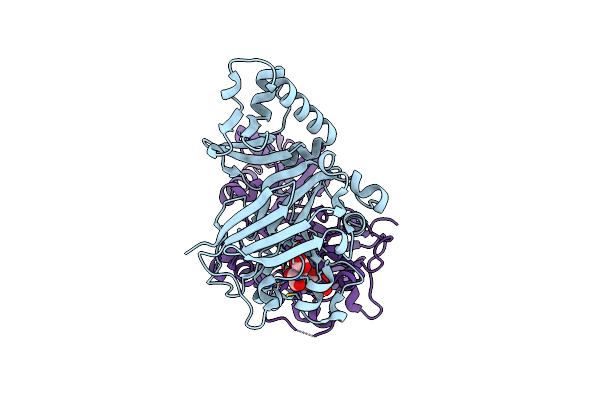 |
Crystal Structure Of Yisk From Bacillus Subtilis In Complex With Reaction Product 4-Hydroxy-2-Oxoglutaric Acid
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: IO9, MN |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: MN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2023-09-20 Classification: HYDROLASE/INHIBITOR Ligands: QBL, GOL |