Search Count: 17
 |
Organism: Bombyx mori
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2024-04-03 Classification: HYDROLASE Ligands: E64 |
 |
Organism: Bombyx mori
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-01-25 Classification: PEPTIDE BINDING PROTEIN |
 |
Crystal Structure Of Juvenile Hormone Acid Methyltransferase Jhamt Mutant Q15E
Organism: Bombyx mori
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2022-04-13 Classification: HORMONE |
 |
Crystal Structure Of Cysteine Proteinase Inhibitor Serpin18 From Bombyx Mori
Organism: Bombyx mori
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2015-09-09 Classification: Hydrolase inhibitor |
 |
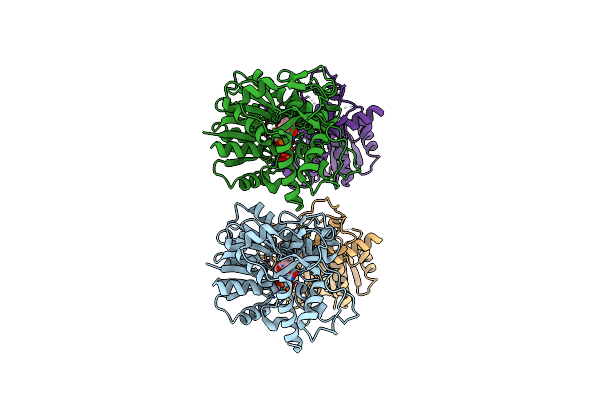
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-10-22 Classification: OXIDOREDUCTASE |
 |
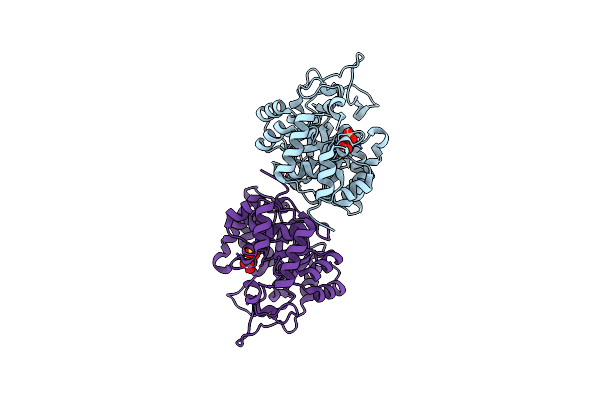
Crystal Structure Of Yeast Methylglyoxal/Isovaleraldehyde Reductase Gre2 Complexed With Nadph
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-10-22 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
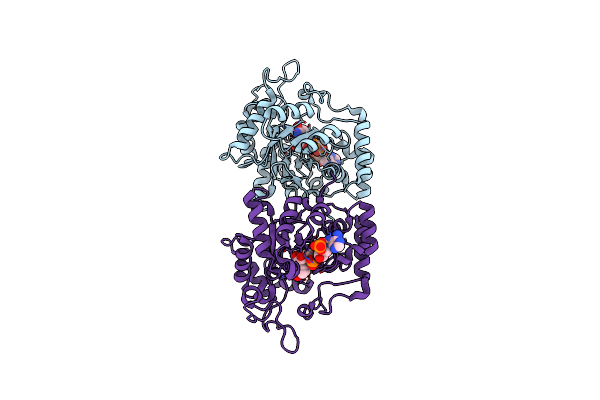
Crystal Structure Of Arabinose Dehydrogenase Ara1 From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-11-27 Classification: OXIDOREDUCTASE Ligands: SO4, GOL |
 |
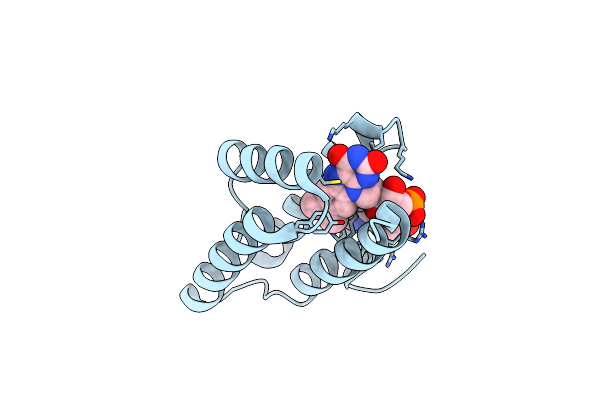
Crystal Structure Of Saccharomyces Cerevisiae Arabinose Dehydrogenase Ara1 Complexed With Nadph
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-11-27 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
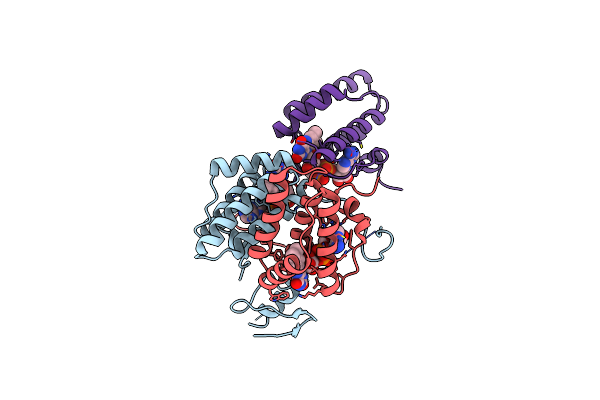
Crystal Structure Of Fad Binding Domain Of Erv1 From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-08-29 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
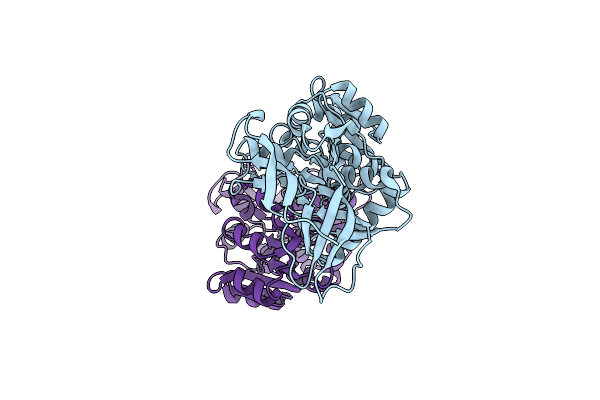
Crystal Structure Of The C30S/C133S Mutant Of Erv1 From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2012-08-29 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
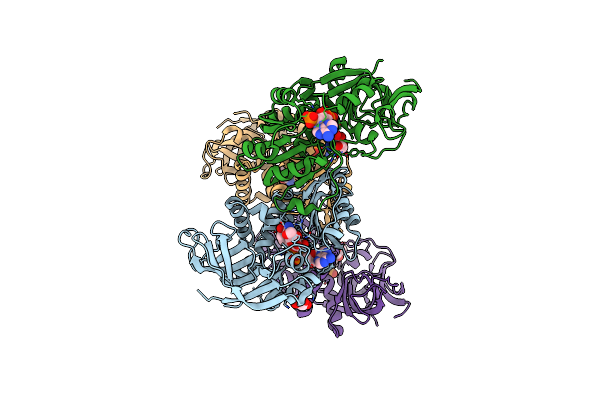
Crystal Structure Of Saccharomyces Cerevisiae Zeta-Crystallin-Like Quinone Oxidoreductase Zta1
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-02-08 Classification: OXIDOREDUCTASE |
 |
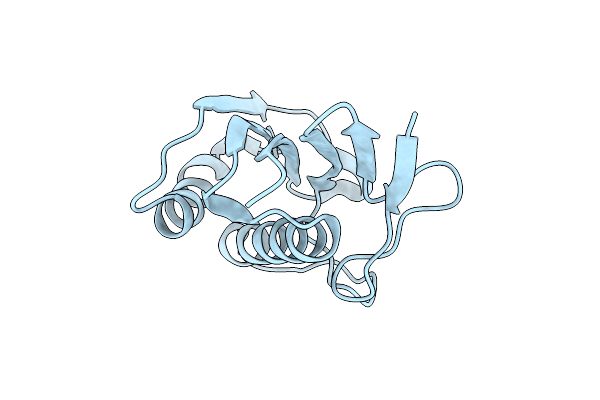
Crystal Structure Of Saccharomyces Cerevisiae Zeta-Crystallin-Like Quinone Oxidoreductase Zta1 Complexed With Nadph
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2012-02-08 Classification: OXIDOREDUCTASE Ligands: NDP, GOL |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2011-06-08 Classification: PROTEIN BINDING |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-06-08 Classification: PROTEIN BINDING |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2011-02-23 Classification: OXIDOREDUCTASE Ligands: ACT |
 |
Crystal Structure Of Mxr1 From Saccharomyces Cerevisiae In Unusual Oxidized Form
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-02-23 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Mxr1 From Saccharomyces Cerevisiae In Complex With Trx2
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2011-02-23 Classification: ELECTRON TRANSPORT/Oxidoreductase |

