Search Count: 281
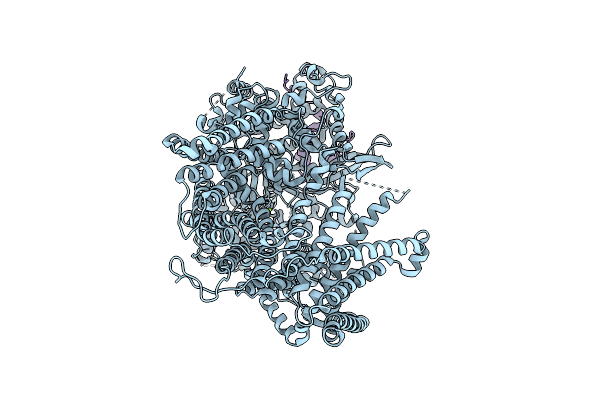 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: MG |
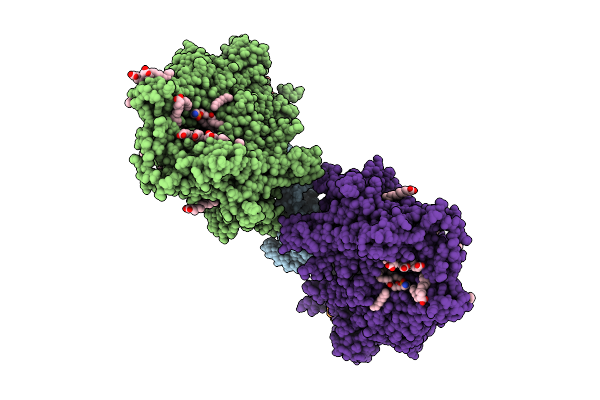 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: MG, DD9, D10, HP6, XKP, PEF, PLM, DCR |
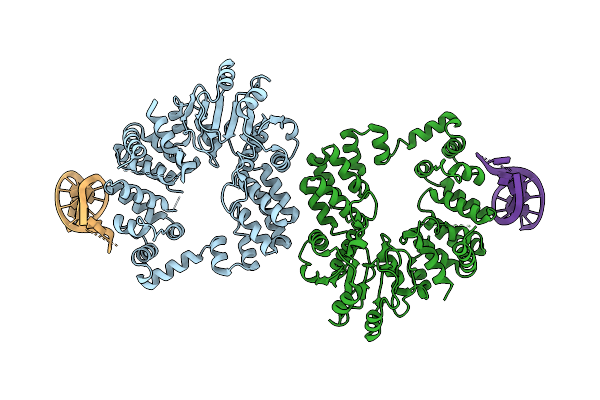 |
Organism: Thermus phage philo
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: RNA BINDING PROTEIN/RNA |
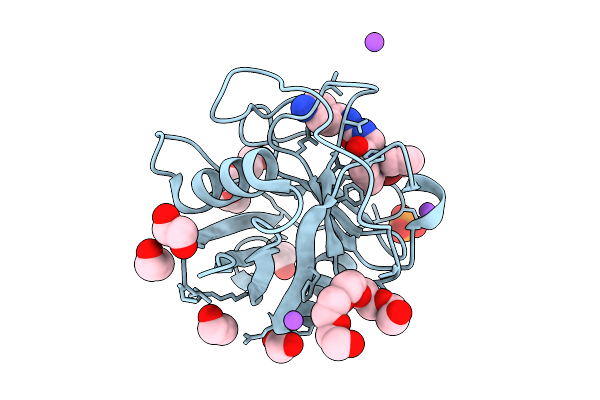 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: ISOMERASE Ligands: 9CK, EDO, 1PE, PGE, PO4, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: ISOMERASE Ligands: A1IU3 |
 |
Organism: Comamonas thiooxydans
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: HYDROLASE Ligands: MLT, SO4, GOL |
 |
Organism: Thermus phage philo
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: RNA BINDING PROTEIN Ligands: CA |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: ISOMERASE Ligands: MN, SO4 |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: METAL BINDING PROTEIN Ligands: MN |
 |
Organism: Orthohantavirus andesense
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: VIRAL PROTEIN |
 |
Organism: Orthohantavirus andesense
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: VIRAL PROTEIN |
 |
Organism: Orthohantavirus andesense
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: VIRAL PROTEIN |
 |
Organism: Orthohantavirus andesense
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: VIRAL PROTEIN |
 |
Organism: Homo sapiens, Orthohantavirus andesense
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Middle east respiratory syndrome-related coronavirus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: VIRAL PROTEIN Ligands: JRY |
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: SUGAR BINDING PROTEIN Ligands: A1IZO, BDF |
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: SUGAR BINDING PROTEIN Ligands: BDF, A1IZO |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: ANTIBIOTIC Ligands: NAG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: ALLERGEN Ligands: NAG |

