Search Count: 29
 |
Organism: Vitis vinifera
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: FE |
 |
Vitis Vinifera Dimeric 13S-Lipoxygenase Loxa In A Detergent Bound Open Conformation
Organism: Vitis vinifera
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: FE, PQE, EDO |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2020-04-29 Classification: DNA BINDING PROTEIN Ligands: SO4, ACE |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2020-04-29 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2020-04-29 Classification: DNA BINDING PROTEIN |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2020-04-29 Classification: DNA BINDING PROTEIN |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719), Chaetomium thermophilum, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-06-14 Classification: TRANSFERASE Ligands: GOL, PO4, CMC |
 |
Organism: Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2017-06-14 Classification: TRANSFERASE |
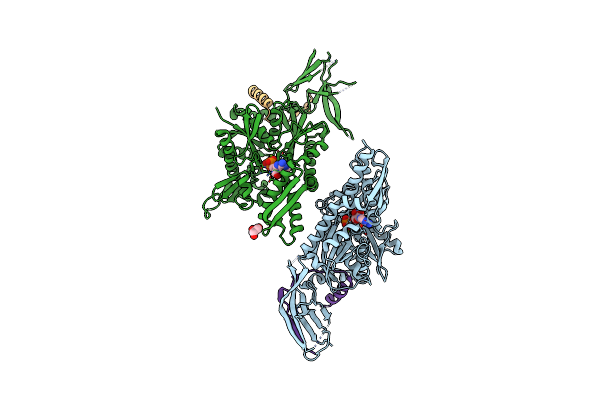 |
Crystal Structure Of The Eukaryotic Ribosome Associated Complex (Rac), A Unique Hsp70/Hsp40 Pair
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719), Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-12-28 Classification: CHAPERONE Ligands: ATP, MG, GOL |
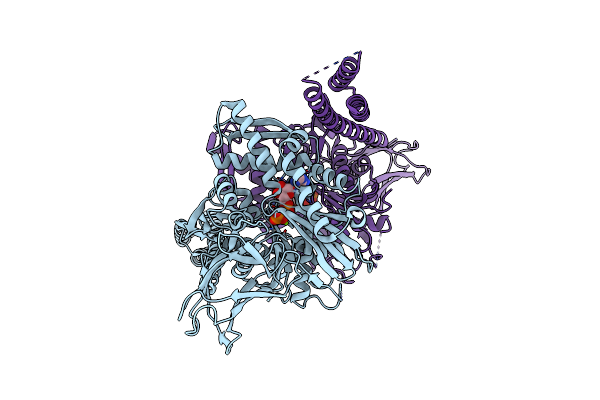 |
Crystal Structure Of The Co-Translational Hsp70 Chaperone Ssb In The Atp-Bound, Open Conformation
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-11-16 Classification: CHAPERONE Ligands: ATP, MG |
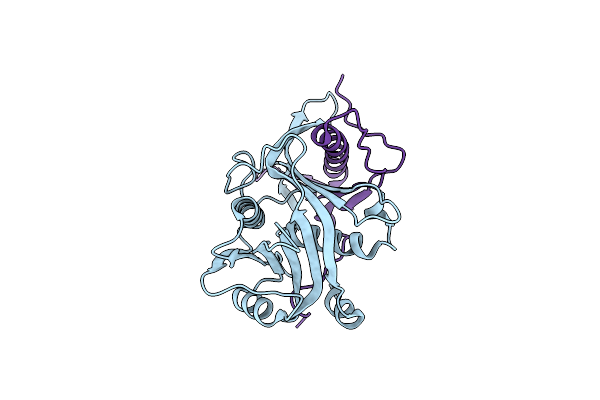 |
Organism: Emericella nidulans fgsc a4
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2015-07-08 Classification: BIOSYNTHETIC PROTEIN |
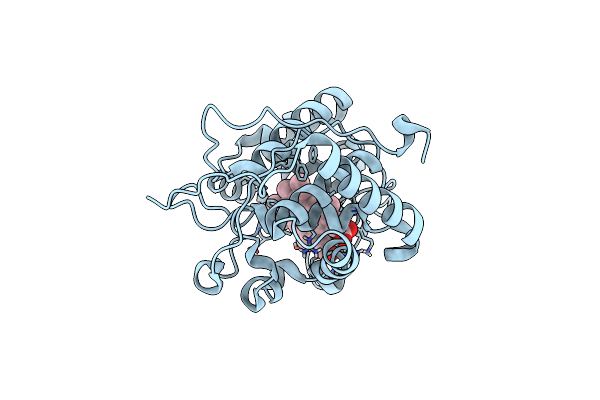 |
Neutron Structure Of Ferric Cytochrome C Peroxidase - Deuterium Exchanged At Room Temperature
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:2.10 Å, 2.407 Å Release Date: 2014-07-16 Classification: OXIDOREDUCTASE Ligands: HEM, DOD |
 |
Neutron Structure Of Compound I Intermediate Of Cytochrome C Peroxidase - Deuterium Exchanged 100 K
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:2.182 Å, 2.501 Å Release Date: 2014-07-16 Classification: OXIDOREDUCTASE Ligands: HEM, DOD |
 |
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2013-08-28 Classification: OXIDOREDUCTASE Ligands: CL, NDP, NO, EDO, HEM, BTB |
 |
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-08-28 Classification: OXIDOREDUCTASE Ligands: CL, NDP, HEM |
 |
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2013-08-28 Classification: OXIDOREDUCTASE Ligands: HEM, NO, CL, NDP, BTB |
 |
Organism: Saccharomyces cerevisiae
Method: NEUTRON DIFFRACTION, X-RAY DIFFRACTION Resolution:2.4000 Å, 2.1000 Å Release Date: 2012-04-04 Classification: OXIDOREDUCTASE Ligands: HEM, DOD |
 |
Organism: Saccharomyces cerevisiae
Method: NEUTRON DIFFRACTION, X-RAY DIFFRACTION Resolution:2.4000 Å, 2.1000 Å Release Date: 2012-04-04 Classification: OXIDOREDUCTASE Ligands: HEM, DOD |
 |
Organism: Glycine max
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-10-12 Classification: OXIDOREDUCTASE Ligands: HEM, SO4 |
 |
Organism: Glycine max
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-10-12 Classification: OXIDOREDUCTASE Ligands: HEM, SO4 |

