Search Count: 69
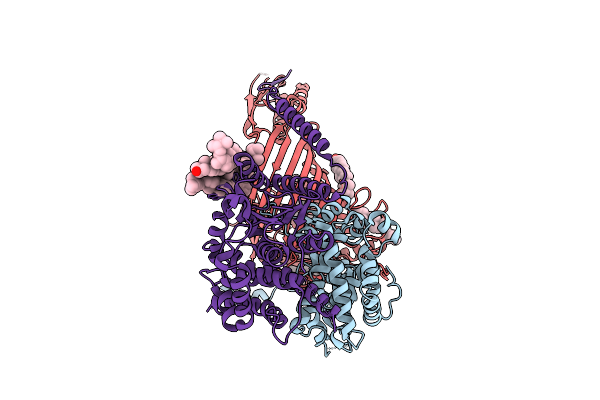 |
Organism: Thermothelomyces thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN Ligands: ERG |
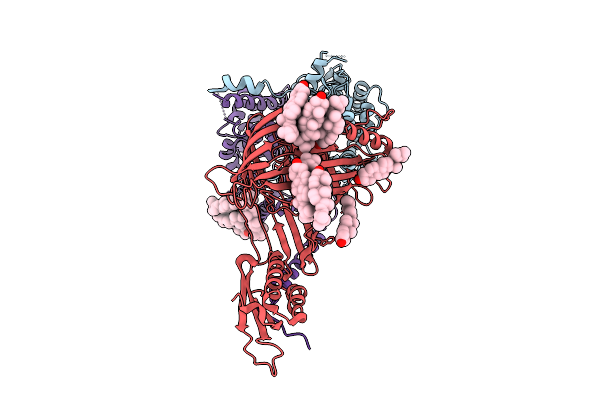 |
Organism: Thermothelomyces thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN Ligands: ERG |
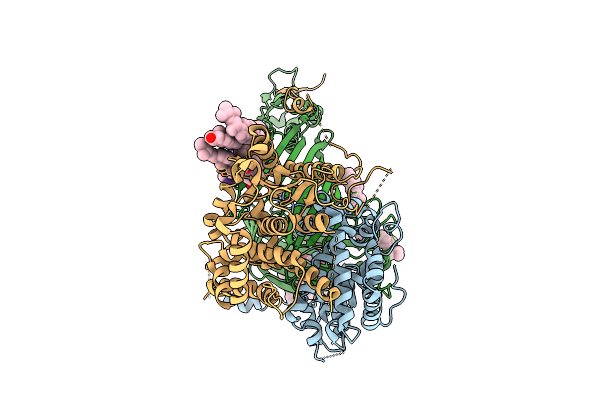 |
Organism: Thermothelomyces thermophilus, Photorhabdus khanii
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN/ANTIBIOTIC Ligands: ERG |
 |
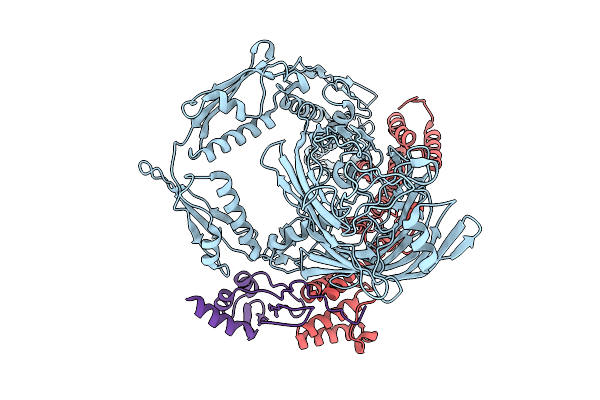
Organism: Neisseria gonorrhoeae fa 1090
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN |
 |
Organism: Neisseria gonorrhoeae
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN |
 |
Organism: Neisseria gonorrhoeae
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-10-30 Classification: PROTEIN TRANSPORT |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2024-10-16 Classification: PROTEIN TRANSPORT |
 |
Organism: Escherichia coli, Escherichia coli 0.1288, Escherichia coli o157
Method: ELECTRON MICROSCOPY Release Date: 2024-05-08 Classification: MEMBRANE PROTEIN/Hydrolase |
 |
Organism: Escherichia coli, Escherichia coli o157
Method: ELECTRON MICROSCOPY Release Date: 2024-05-08 Classification: MEMBRANE PROTEIN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-05-08 Classification: MEMBRANE PROTEIN |
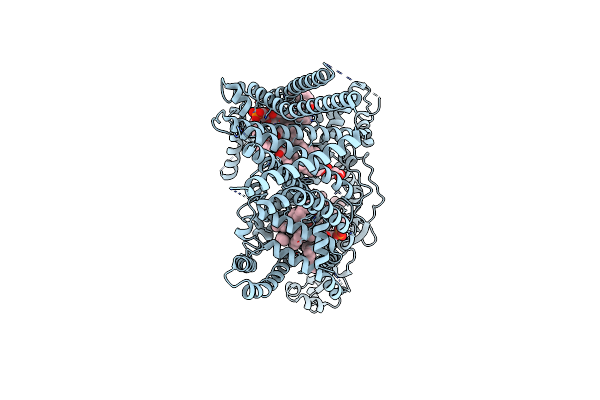 |
Cryo-Em Structure Of E3 Ubiquitin Ligase Doa10 From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae by4741
Method: ELECTRON MICROSCOPY Release Date: 2024-03-13 Classification: LIGASE Ligands: PC1, TGL, Y01, ERG |
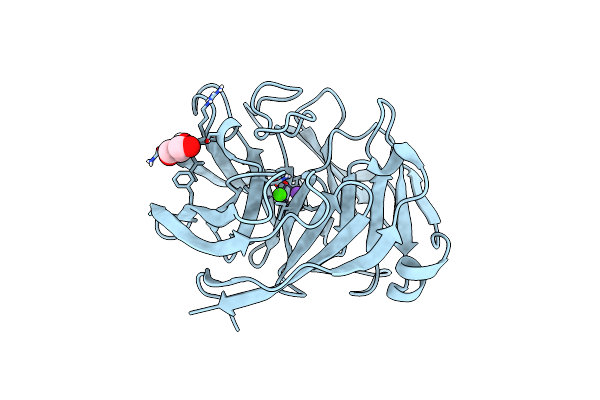 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2024-01-17 Classification: PROTEIN BINDING Ligands: GOL, CA, NA |
 |
Organism: Marine sediment metagenome
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2023-09-13 Classification: ANTIFREEZE PROTEIN Ligands: PG4 |
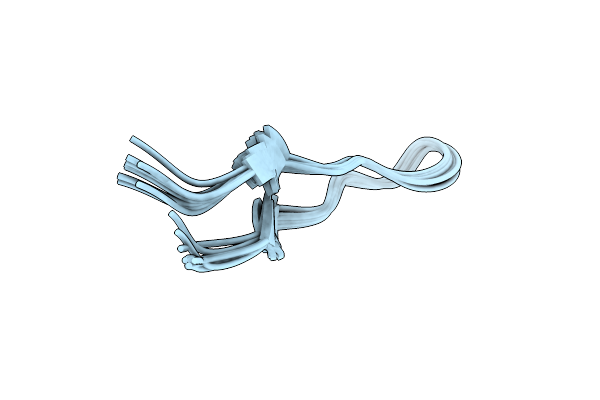 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2023-09-06 Classification: DE NOVO PROTEIN |
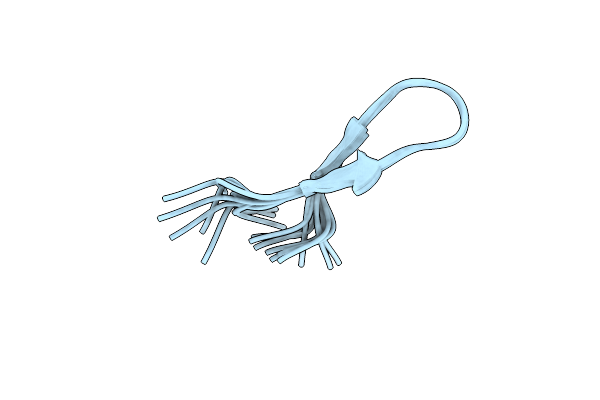 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2023-06-28 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2023-06-28 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2023-06-28 Classification: DE NOVO PROTEIN |
 |
Organism: Mus musculus, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-06-21 Classification: TRANSLOCASE/IMMUNE SYSTEM Ligands: CDL, PTY |
 |
Organism: Saccharomyces cerevisiae, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-06-21 Classification: TRANSLOCASE Ligands: CDL, PTY |

