Search Count: 18
 |
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2025-05-14 Classification: PLANT PROTEIN Ligands: GOL, ETE, MAN |
 |
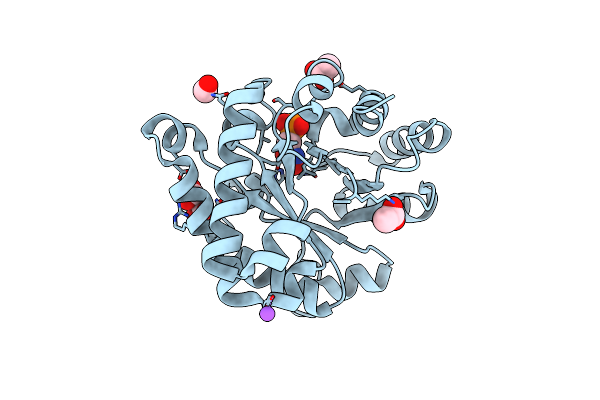
Complex Between The Porcine Trypsin And M271 A Kunitz-Sti From Solanum Tuberosum
Organism: Solanum tuberosum, Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2025-05-14 Classification: HYDROLASE Ligands: CA |
 |
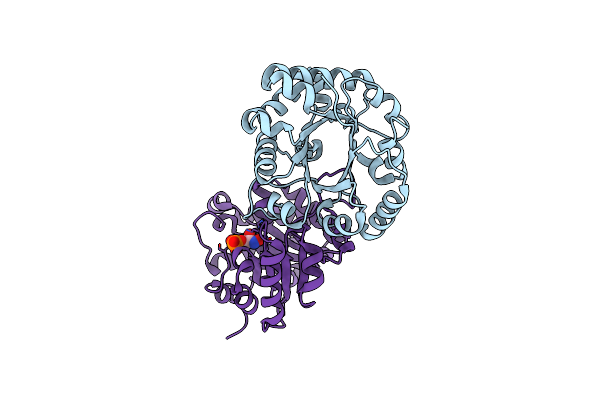
Crystal Structure Of The Reconstruction Of The Ancestral Triosephosphate Isomerase Of The Last Opisthokont Common Ancestor Obtained By Bayesian Inference With Pgh
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2024-09-04 Classification: ISOMERASE |
 |
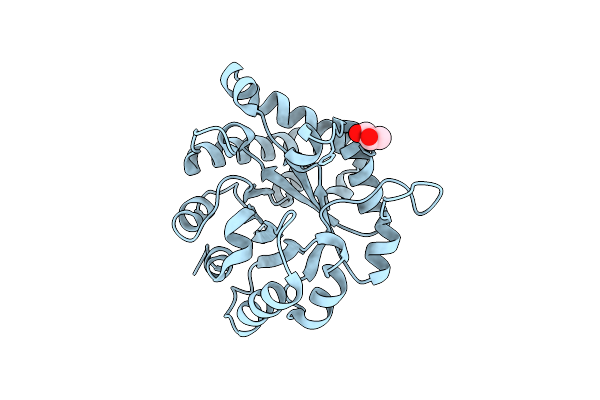
Crystal Structure Of The Worst Case Of The Reconstruction Of The Ancestral Triosephosphate Isomerase Of The Last Opisthokont Common Ancestor Obtained By Maximum Likelihood With Pgh
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-09-04 Classification: ISOMERASE Ligands: PGH |
 |
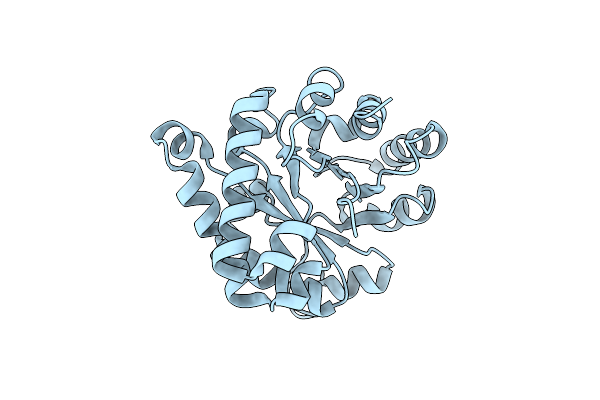
Crystal Structure Of The Ancestral Triosephosphate Isomerase Reconstruction Of The Last Opisthokont Common Ancestor Obtained By Bayesian Inference
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2024-09-04 Classification: ISOMERASE Ligands: GOL |
 |
Crystal Structure Of The Reconstruction Of The Worst Case Of The Ancestral Triosephosphate Isomerase Of The Last Opisthokont Common Ancestor Obtained By Bayesian Inference
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2024-09-04 Classification: ISOMERASE |
 |
Crystal Structure Of The Reconstruction Of The Ancestral Triosephosphate Isomerase Of The Last Opisthokont Common Ancestor Obtained By Maximum Likelihood
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2024-09-04 Classification: ISOMERASE |
 |
Crystal Structure Of The Reconstruction Of The Ancestral Triosephosphate Isomerase Of The Last Opisthokont Common Ancestor Obtained By Maximum Likelihood With Pgh
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2024-09-04 Classification: ISOMERASE Ligands: PGH, ACY, PGE, ACT, NA, EDO, PEG, CL |
 |
Crystal Structure Of The Worst Case Reconstruction Of The Ancestral Triosephosphate Isomerase Of The Last Opisthokont Common Ancestor Obtained By Maximum Likelihood
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2024-09-04 Classification: ISOMERASE Ligands: F |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2021-07-21 Classification: DE NOVO PROTEIN Ligands: GOL, CL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2021-07-21 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2021-07-21 Classification: DE NOVO PROTEIN |
 |
Crystal Structure At Ph 7.0 Of A Potato Sti-Kunitz Bi-Functional Inhibitor Of Serine And Aspartic Proteases In Space Group P4322 And Ph 9.0
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2016-07-06 Classification: HYDROLASE INHIBITOR |
 |
Crystal Structure At Ph 9.0 Of A Potato Sti-Kunitz Bi-Functional Inhibitor Of Serine And Aspartic Proteases In Space Group P4322 And Ph 9.0
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2016-07-06 Classification: HYDROLASE INHIBITOR |
 |
Crystal Structure Of N19D Potato Sti-Kunitz Bi-Functional Inhibitor Of Serine And Aspartic Proteases In Space Group P4322 And Ph 7.2
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2016-07-06 Classification: HYDROLASE INHIBITOR |
 |
Crystal Structure Of N19D Potato Sti-Kunitz Bi-Functional Inhibitor Of Serine And Aspartic Proteases In Space Group C2221 And Ph 3.5
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2016-07-06 Classification: HYDROLASE INHIBITOR |
 |
Crystal Structure Of Potato Sti-Kunitz Bi-Functional Inhibitor Of Serine And Aspartic Proteases In Space Group P22121 And Ph 7.0
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2016-07-06 Classification: HYDROLASE |
 |
Crystal Structure Of A Potato Sti-Kunitz Bifunctional Inhibitor Of Serine And Aspartic Proteases In Space Group P4322 And Ph 7.4
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-07-06 Classification: HYDROLASE INHIBITOR Ligands: NAG |

