Search Count: 359
 |
Coupling Of Polymerase-Nucleoprotein-Rna In An Influenza Virus Mini Ribonucleoprotein Complex
Organism: Influenza a virus (a/victoria/3/1975(h3n2))
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: VIRAL PROTEIN/RNA |
 |
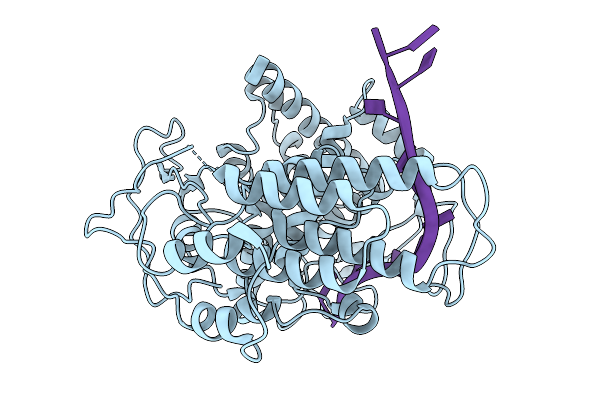
Coupling Of Polymerase-Nucleoprotein-Rna In An Influenza Virus Mini Ribonucleoprotein Complex
Organism: Influenza a virus (a/victoria/3/1975(h3n2))
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: VIRAL PROTEIN/RNA |
 |
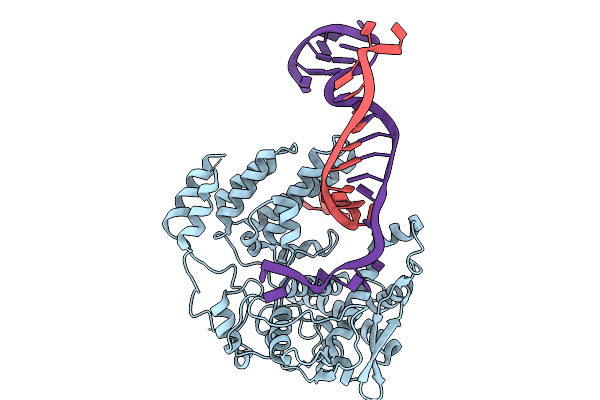
Coupling Of Polymerase-Nucleoprotein-Rna In An Influenza Virus Mini Ribonucleoprotein Complex
Organism: Influenza a virus (a/victoria/3/1975(h3n2))
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: VIRAL PROTEIN/RNA |
 |
Coupling Of Polymerase-Nucleoprotein-Rna In An Influenza Virus Mini Ribonucleoprotein Complex
Organism: Influenza a virus (a/victoria/3/1975(h3n2))
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: VIRAL PROTEIN/RNA |
 |
Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Drtc) In Post-Capping State
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: VIRAL PROTEIN/RNA Ligands: ZN |
 |
Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) In Post-Capping State
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: VIRAL PROTEIN/RNA Ligands: ZN |
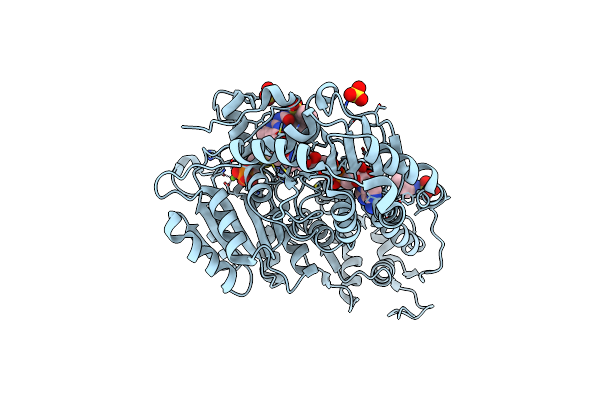 |
Crystal Structure Of Arabidopsis Thaliana Acetohydroxyacid Synthase In Complex With 2022-Ls5
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: TRANSFERASE Ligands: MG, A1AU7, FAD, AUJ, NHE, SO4 |
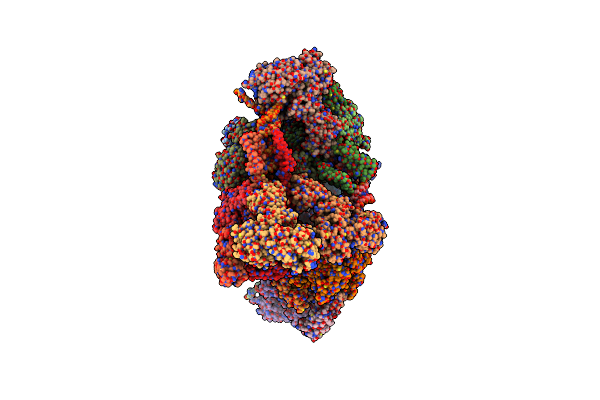 |
Sars-Cov-2 Replication-Transcription Complex Has A Dimer-Of-Dimeric Architecture (Ddrtc) In Pre-Capping Initiation.
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: VIRAL PROTEIN Ligands: ZN, ADP, MG, PO4 |
 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Atp|Adp-Bound Ifasym-2 State
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ATP, MG, ADP |
 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Adp-Bound Ifasym-3 State (Atp 37 Degrees C Treated
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ADP, MG |
 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Adp-Bound Ifasym-3 State (Adp 4 Degrees C Treated)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ADP, MG |
 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Adp-Bound Ifasym-3 (Peptidisc) State (Atp 37Degrees C Treated)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ADP |
 |
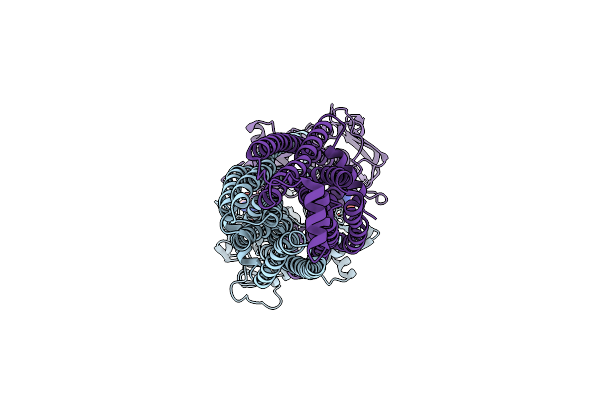
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Adp-Bound Ifasym-3 (Peptidisc) State (Adp 4Degrees C Treated)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ADP |
 |
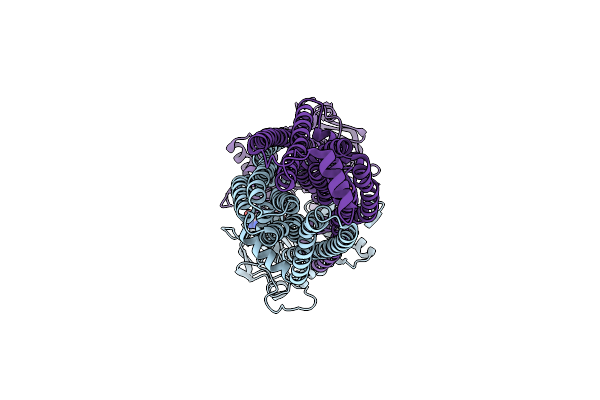
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Atp|Adp+Vi-Bound Occ (Vi) State
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ATP, MG, ADP, VO4 |
 |
Cryo-Em Structure Of Msrv1273C/72C(E553Q) Mutant From Mycobacterium Smegmatis In The Atp-Bound Occ State
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ATP, MG |
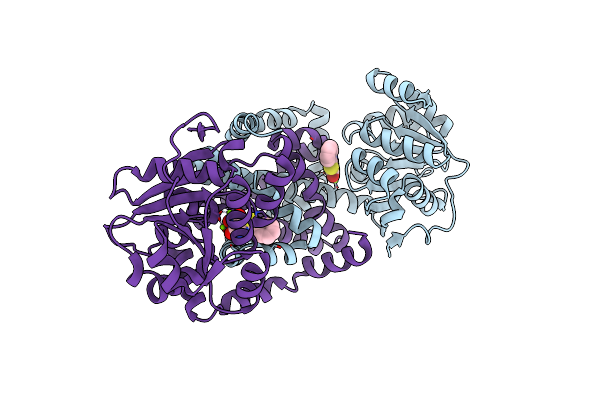 |
Crystal Structure Of Staphylococcus Aureus Ketol-Acid Reductoisomerase In Complex With Mg2+, And Jk-5-114
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-04-09 Classification: ISOMERASE Ligands: A1AKJ, MG |
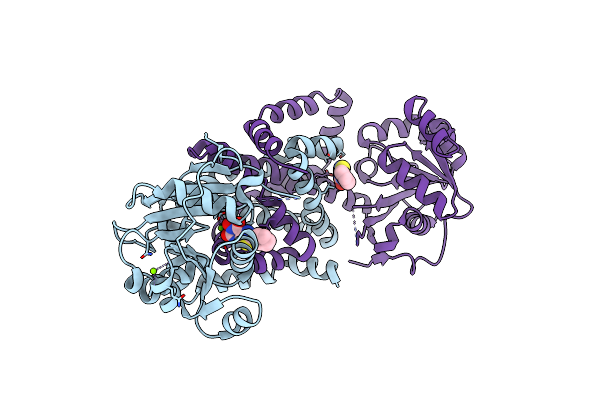 |
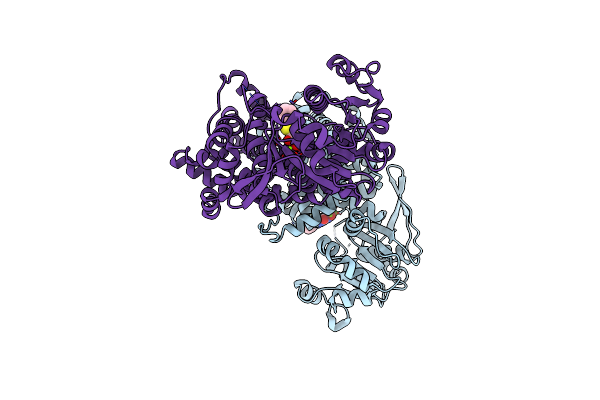
Crystal Structure Of Staphylococcus Aureus Ketol-Acid Reductoisomerase In Complex With Mg2+, And Jk-5-115
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-04-09 Classification: ISOMERASE Ligands: MG, A1AKY |
 |
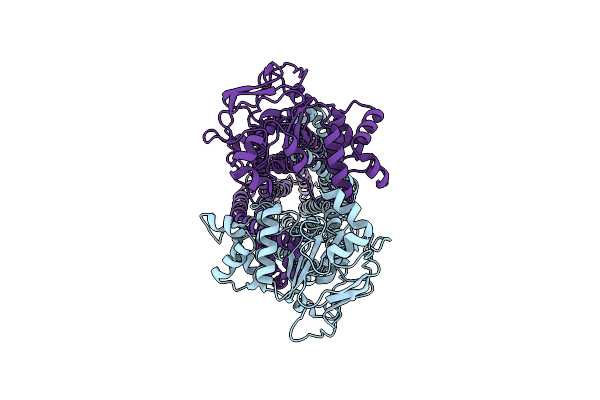
Crystal Structure Of Oryza Sativa Ketol-Acid Reductoisomerase In Complex With Mg2+, And Jk-5-114
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Release Date: 2025-04-09 Classification: ISOMERASE Ligands: MG, A1AKJ |
 |
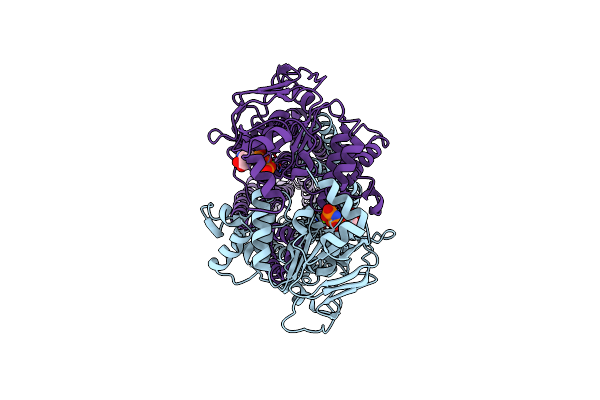
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Ifapo State
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: TRANSPORT PROTEIN |
 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Amppnp-Bound Ifasym-1 State
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: TRANSPORT PROTEIN Ligands: ANP, MG |

