Search Count: 69
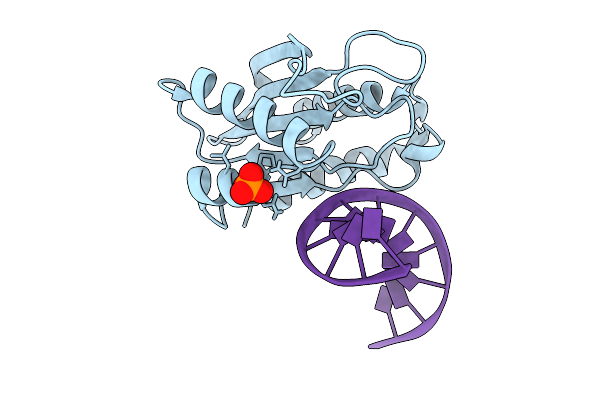 |
Organism: Thiopseudomonas alkaliphila, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: DNA BINDING PROTEIN/DNA Ligands: PO4 |
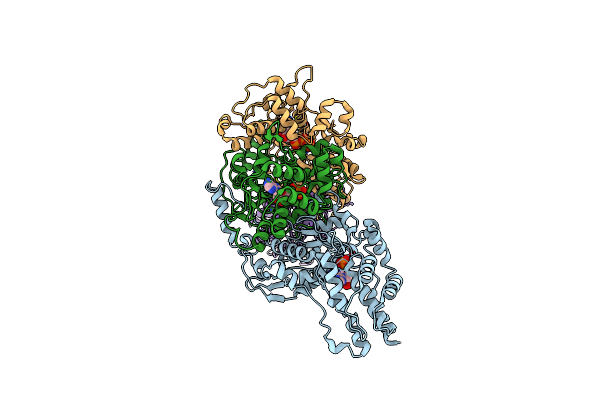 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: DNA BINDING PROTEIN Ligands: ATP, MG, ZN, ADP |
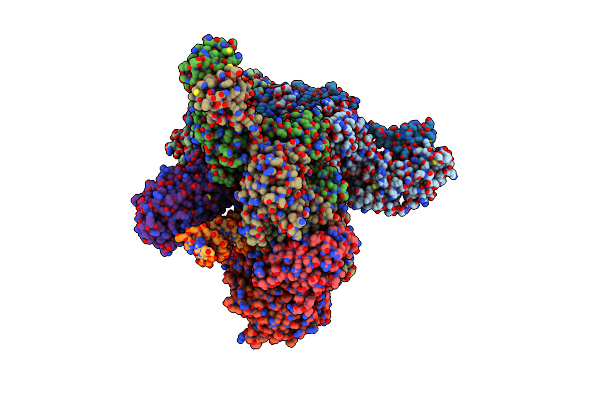 |
Cryoem Structure Of The Tnsc(1-503)-Tnsd(1-318)-Dna Complex In A 7:2:1 Stoichiometry From E. Coli Tn7
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: DNA BINDING PROTEIN/DNA Ligands: ADP, MG, ATP, ZN |
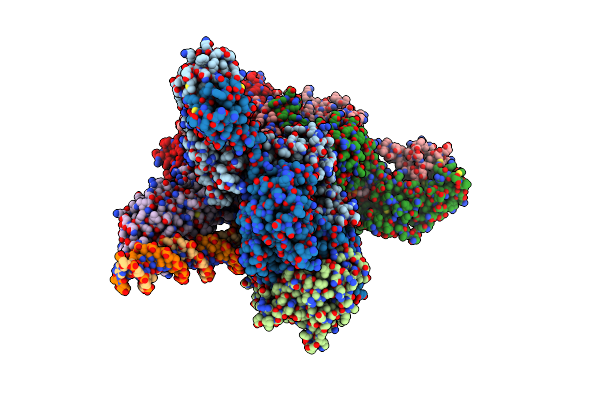 |
Cryoem Structure Of The Tnsc(1-503)-Tnsd(1-318)-Dna Complex In A 6:2:1 Stoichiometry From E. Coli Tn7
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: DNA BINDING PROTEIN/DNA Ligands: ATP, MG, ZN, ADP |
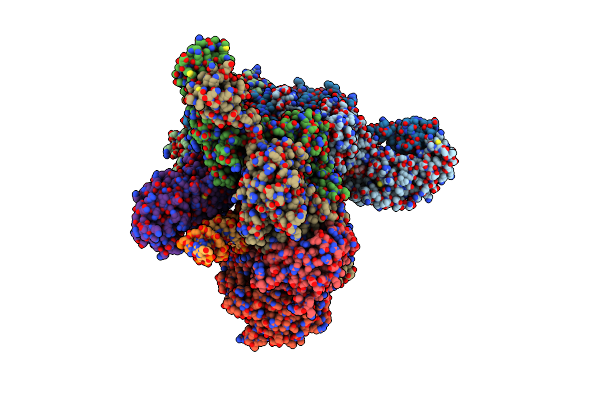 |
Cryoem Structure Of The Tnsc(1-503)-Tnsd(1-318)-Dna Complex In A 7:2:1 Stoichiometry From E. Coli Tn7 Bound To Atpgs And Adp
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: DNA BINDING PROTEIN/DNA Ligands: ADP, MG, AGS, ZN |
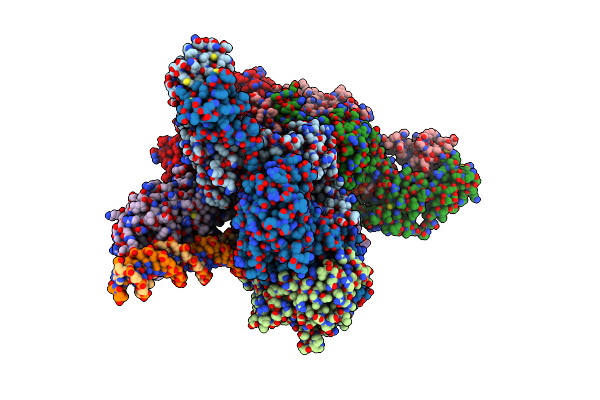 |
Cyoem Structure Of The Tnsc(1-503)-Tnsd(1-318)-Dna Complex In A 6:2:1 Stoichiometry From E. Coli Tn7 Bound To Atpgs And Adp
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: DNA BINDING PROTEIN/DNA Ligands: AGS, MG, ZN, ADP |
 |
Organism: Trypanosoma cruzi
Method: SOLUTION NMR Release Date: 2024-05-08 Classification: LIPID BINDING PROTEIN |
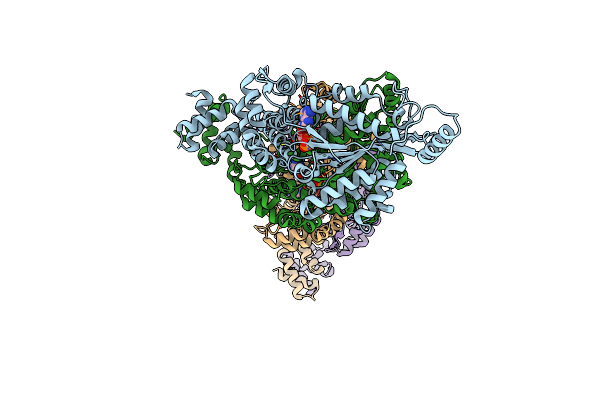 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2022-02-23 Classification: DNA BINDING PROTEIN Ligands: ADP, MG |
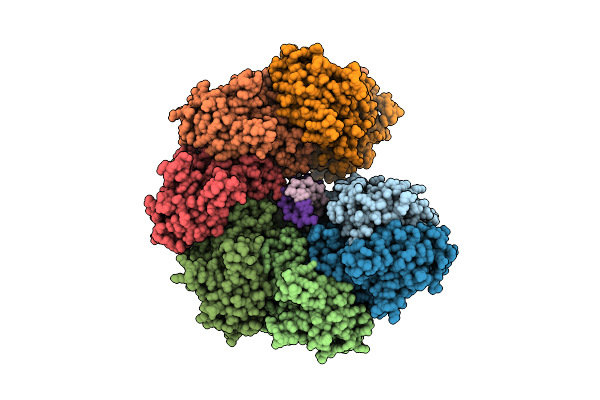 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-02-23 Classification: DNA BINDING PROTEIN/DNA Ligands: ANP, MG |
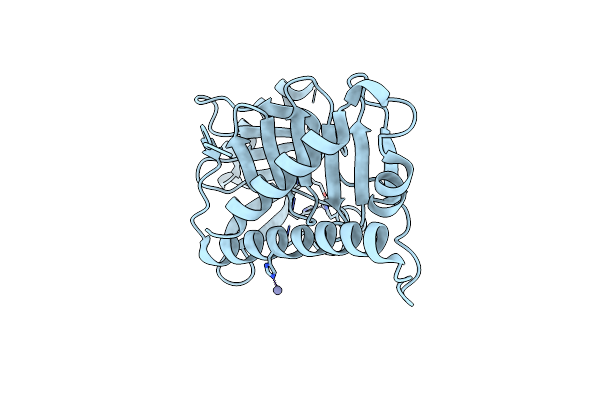 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-02-19 Classification: CELL CYCLE, Transferase Ligands: ZN |
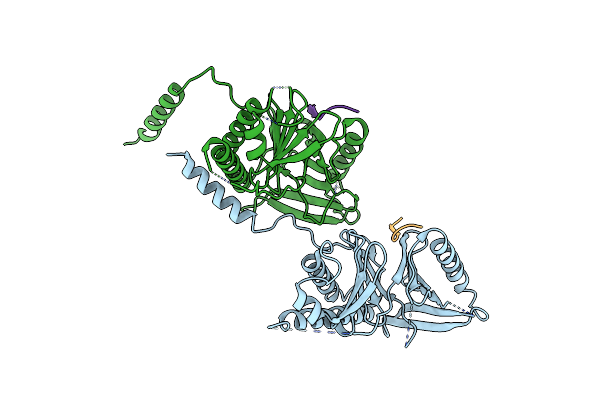 |
Crystal Structure Of Budding Yeast Cdc5 Polo-Box Domain In Complex With Spc72 Phosphopeptide.
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-02-19 Classification: CELL CYCLE, Transferase Ligands: CL |
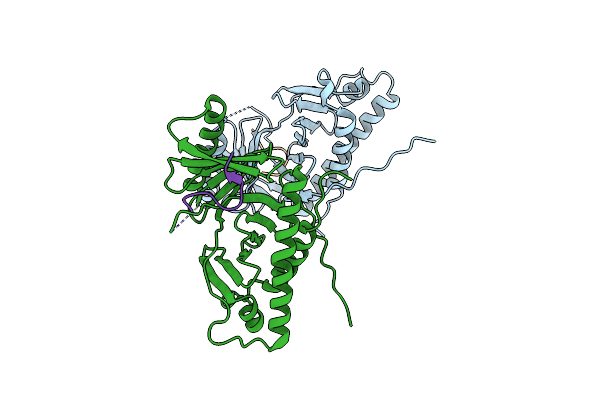 |
Crystal Structure Of Budding Yeast Cdc5 Polo-Box Domain In Complex With The Dbf4 Polo-Interacting Region.
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2020-02-19 Classification: CELL CYCLE, Transferase |
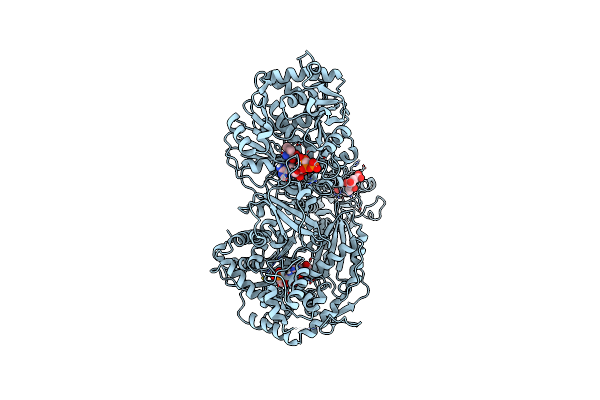 |
Crystal Structure Of A 4-Domain Construct Of Lgra In The Substrate Donation State
Organism: Brevibacillus parabrevis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-11-20 Classification: LIGASE Ligands: JQG, FON, APC, VAL, PO4, MG |
 |
Crystal Structure Of A 4-Domain Construct Of A Mutant Of Lgra In The Substrate Donation State
Organism: Brevibacillus parabrevis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-11-20 Classification: LIGASE Ligands: 9EF, APC, VAL |
 |
Crystal Structure Of A 5-Domain Construct Of Lgra In The Substrate Donation State
Organism: Brevibacillus parabrevis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-11-20 Classification: LIGASE Ligands: PNS, PO4 |
 |
Organism: Brevibacillus parabrevis
Method: X-RAY DIFFRACTION Resolution:6.00 Å Release Date: 2019-11-20 Classification: LIGASE Ligands: PNS |
 |
Organism: Brevibacillus parabrevis
Method: X-RAY DIFFRACTION Resolution:6.00 Å Release Date: 2019-11-20 Classification: LIGASE Ligands: DG9 |
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2019-09-18 Classification: RIBOSOME |
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2019-09-18 Classification: RIBOSOME Ligands: GNP |
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2019-09-18 Classification: RIBOSOME |

