Search Count: 27
 |
Cryo-Em Structure Of The Human Sk2-4 Chimera/Calmodulin Channel Complex In The Ca2+ Bound State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: TRANSPORT PROTEIN Ligands: K, CA |
 |
Cryo-Em Structure Of The Human Sk2-4 Chimera/Calmodulin Channel Complex In The Ca2+ Free State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: TRANSPORT PROTEIN Ligands: K, CA |
 |
Cryo-Em Structure Of The Human Sk2-4 Chimera/Calmodulin Channel Complex Bound To The Bee Toxin Apamin
Organism: Homo sapiens, Apis mellifera
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: TRANSPORT PROTEIN/TOXIN Ligands: K, CA |
 |
Cryo-Em Structure Of The Human Sk2-4 Chimera/Calmodulin Channel Complex Bound To A Small Molecule Inhibitor
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: TRANSPORT PROTEIN/INHIBITOR Ligands: A1B8D, K, CA |
 |
Cryo-Em Structure Of The Human Sk2-4 Chimera/Calmodulin Channel Complex Bound To A Small Molecule Activator
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: TRANSPORT PROTEIN Ligands: A1B8G, K, CA |
 |
Cryo-Em Structure Of Conformation 1 Of Complex Of Nipah Virus Attachment Glycoprotein G With 1E5 Neutralizing Antibody
Organism: Macaca mulatta, Nipah virus
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: VIRAL PROTEIN |
 |
Cryo-Em Structure Of Conformation 2 Of Complex Of Nipah Virus Attachment G With 1E5 Neutralizing Antibody
Organism: Macaca mulatta, Nipah virus
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: VIRAL PROTEIN |
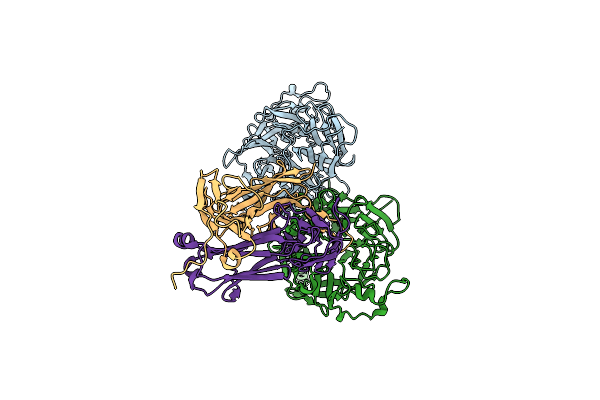 |
Organism: Homo sapiens, Henipavirus nipahense
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Nipah Virus Attachment Glycoprotein Head Domain In Complex With A Broadly Neutralizing Antibody 1E5
Organism: Henipavirus nipahense, Macaca mulatta
Method: X-RAY DIFFRACTION Resolution:3.24 Å Release Date: 2024-01-24 Classification: ANTIVIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
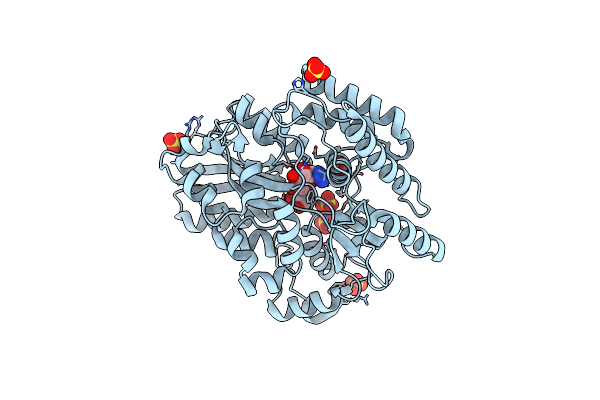 |
Crystal Structure Of Mycobacterium Tuberculosis Lpqy In Complex With Trehalose Analogue Yb-03
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-10-04 Classification: SUGAR BINDING PROTEIN Ligands: SO4 |
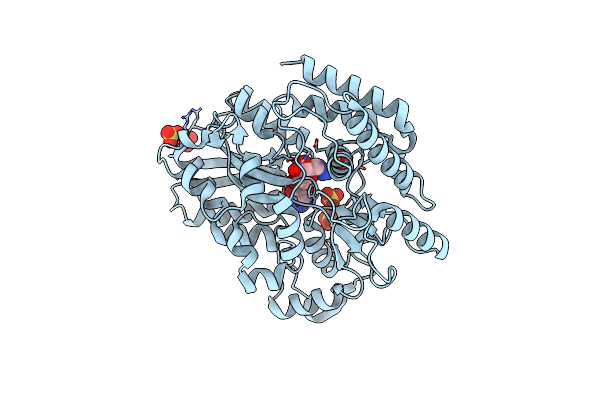 |
Crystal Structure Of Mycobacterium Tuberculosis Lpqy In Complex With Trehalose Analogue Yb-04
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-10-04 Classification: SUGAR BINDING PROTEIN Ligands: SO4, TY6 |
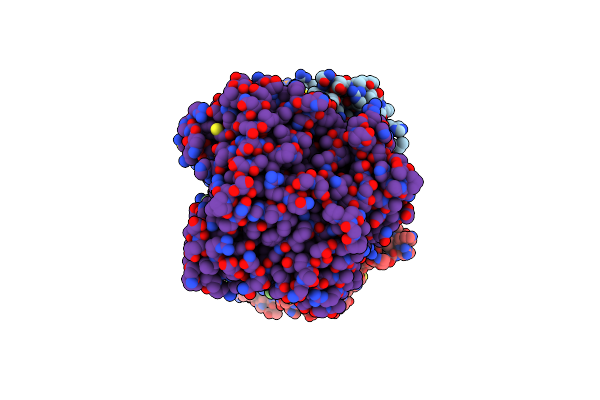 |
Cryo-Em Structure Of Mycobacterium Tuberculosis Lpqy-Sugabc In Complex With Trehalose
Organism: Mycobacterium tuberculosis h37rv
Method: ELECTRON MICROSCOPY Release Date: 2023-09-27 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of Mycobacterium Tuberculosis Lpqy With Trehalose Bound In A Closed Liganded Form
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-09-27 Classification: SUGAR BINDING PROTEIN Ligands: SO4 |
 |
Crystal Structure Of Mycobacterium Tuberculosis Lpqy In Complex With Trehalose Analogue Yb-06
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-09-27 Classification: SUGAR BINDING PROTEIN Ligands: U0X, SO4 |
 |
Crystal Structure Of Mycobacterium Tuberculosis Lpqy In Complex With Trehalose Analogue Yb-16
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-09-27 Classification: SUGAR BINDING PROTEIN Ligands: SO4, W4Z, GLC |
 |
Crystal Structure Of Mycobacterium Tuberculosis Lpqy In Complex With Trehalose Analogue Yb-17
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-09-27 Classification: SUGAR BINDING PROTEIN Ligands: BEZ, SO4 |
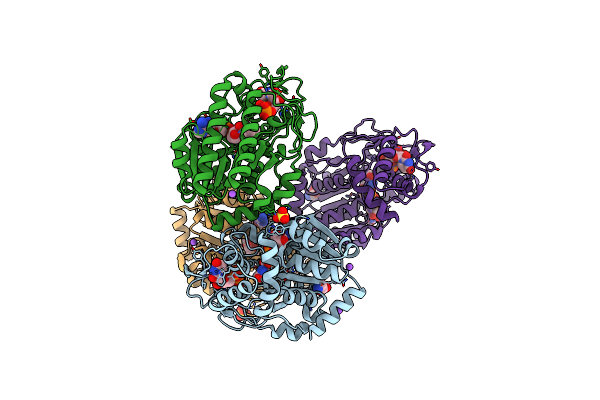 |
Specific Recognition Of N-Acetylated Substrates And Domain Flexibility In Wbgu: A Udp-Galnac 4-Epimerase
Organism: Plesiomonas shigelloides
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2011-05-25 Classification: ISOMERASE Ligands: NAD, GLY, NA, UNL, SO4 |
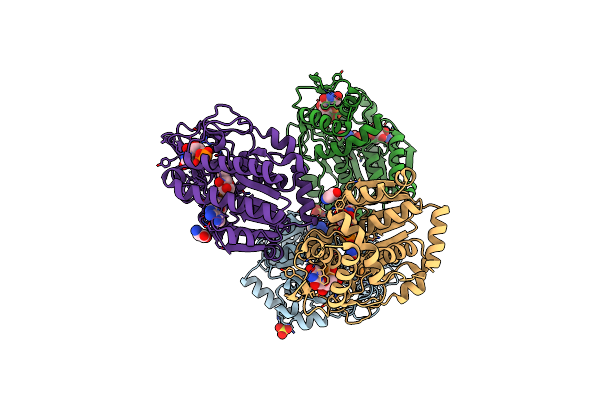 |
Specific Recognition Of N-Acetylated Substrates And Domain Flexibility In Wbgu: A Udp-Galnac 4-Epimerase
Organism: Plesiomonas shigelloides
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2011-05-25 Classification: ISOMERASE Ligands: NAD, SO4, UNL, GLY |
 |
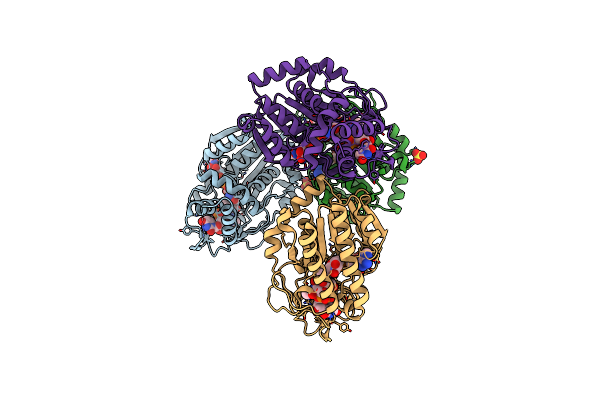
Specific Recognition Of N-Acetylated Substrates And Domain Flexibility In Wbgu: A Udp-Galnac 4-Epimerase
Organism: Plesiomonas shigelloides
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-05-25 Classification: ISOMERASE Ligands: NAD, UNL, SO4 |
 |
Specific Recognition Of N-Acetylated Substrates And Domain Flexibility In Wbgu: A Udp-Galnac 4-Epimerase
Organism: Plesiomonas shigelloides
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-05-25 Classification: ISOMERASE Ligands: NAD, UD2, SO4 |

