Search Count: 88
 |
Organism: Marinobacter sp. dsm 11874
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: 1GP |
 |
Organism: Marinobacter sp. dsm 11874
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: G3P |
 |
Organism: Phaeobacter sp. med193
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: 1GP |
 |
Organism: Phaeobacter sp. med193
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: G3P |
 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: PROTEIN TRANSPORT |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: PROTEIN TRANSPORT |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: PROTEIN TRANSPORT |
 |
Organism: Listeria monocytogenes
Method: ELECTRON MICROSCOPY Release Date: 2025-01-08 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Listeria monocytogenes
Method: ELECTRON MICROSCOPY Release Date: 2025-01-08 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Listeria monocytogenes
Method: ELECTRON MICROSCOPY Release Date: 2025-01-08 Classification: VIRUS LIKE PARTICLE Ligands: FE |
 |
Organism: Myroides profundi
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: ELECTRON TRANSPORT |
 |
Organism: Myroides profundi
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: ELECTRON TRANSPORT Ligands: KEN |
 |
Organism: Myroides profundi
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: ELECTRON TRANSPORT Ligands: KEN |
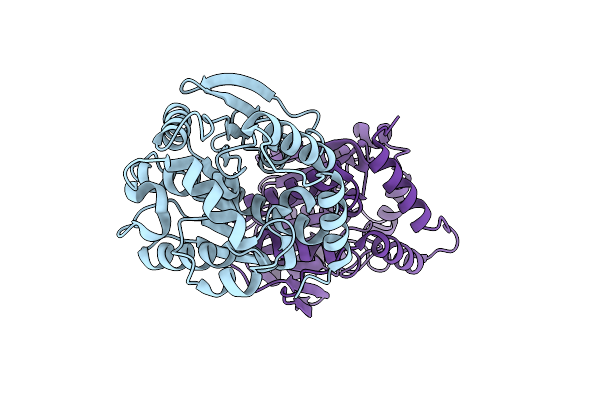 |
Organism: Paracoccus kondratievae
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2024-07-31 Classification: HYDROLASE |
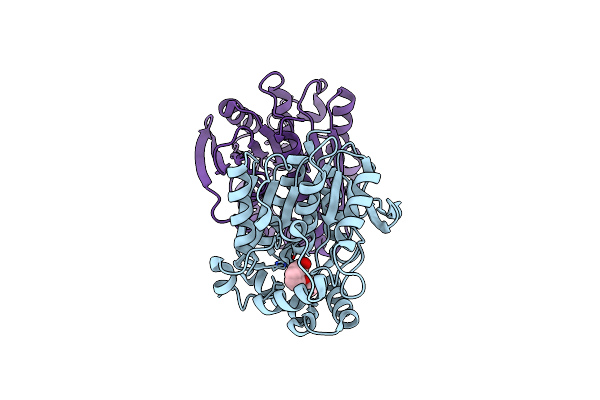 |
Organism: Paracoccus kondratievae
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: A1D6Y |
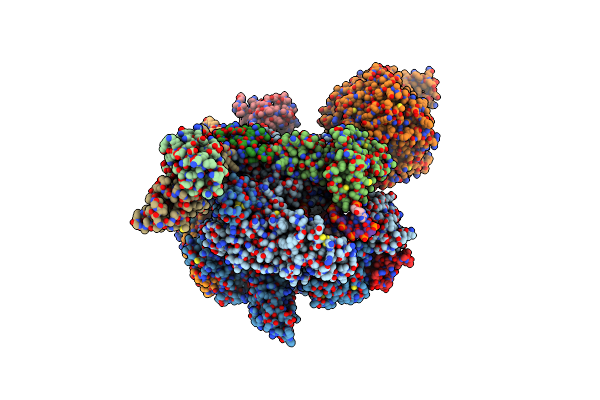 |
Organism: Arabidopsis thaliana, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: TRANSCRIPTION/DNA/RNA Ligands: ZN, MG, GTP |
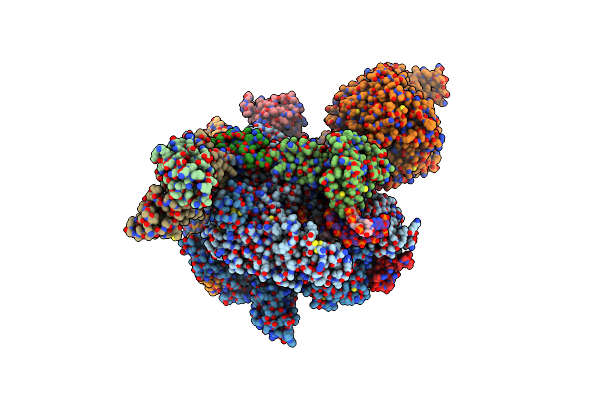 |
Organism: Arabidopsis thaliana, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: TRANSCRIPTION/DNA/RNA Ligands: ZN, MG |
 |
Organism: Arabidopsis thaliana, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: TRANSCRIPTION/DNA/RNA Ligands: ZN, MG |
 |
Organism: Arabidopsis thaliana, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: TRANSCRIPTION/DNA/RNA Ligands: ZN, MG |
 |
Organism: Arabidopsis thaliana, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: STRUCTURAL PROTEIN Ligands: BEF, ADP |

