Search Count: 25
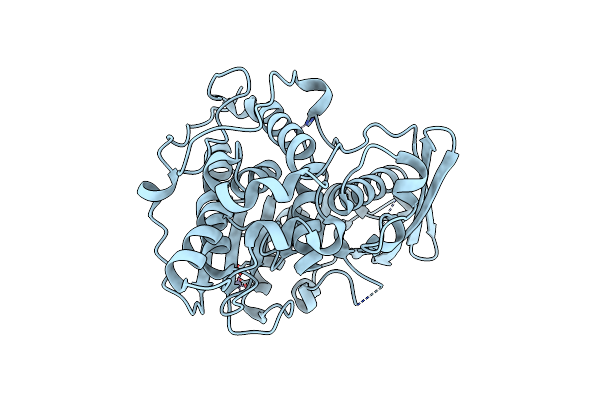 |
Crystal Structure Of A Ce15 From Fibrobacter Succinogenes Subsp. Succinogenes S85
Organism: Fibrobacter succinogenes subsp. succinogenes
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-08-28 Classification: HYDROLASE Ligands: ZN |
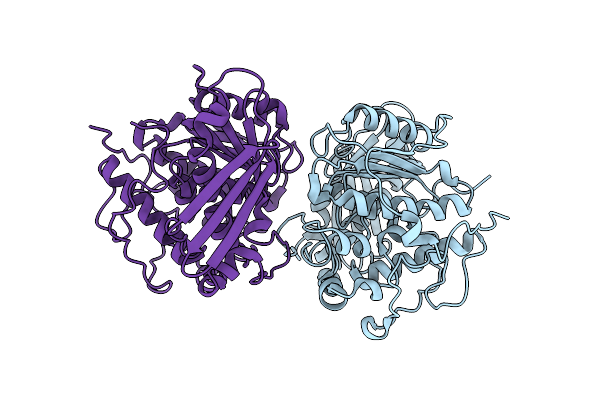 |
Crystal Structure Of A Ce15 Glucuronoyl Esterase From Piromyces Rhizinflatus
Organism: Piromyces rhizinflatus
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2024-08-28 Classification: HYDROLASE |
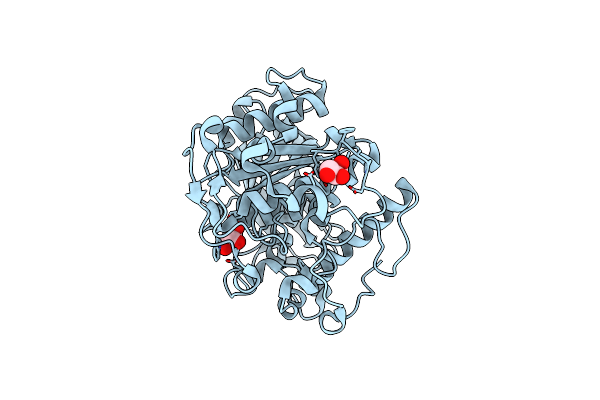 |
Crystal Structure Of A Ce15 Glucuronoyl Esterase From Ruminococcus Flavefaciens
Organism: Ruminococcus flavefaciens 17
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2024-08-28 Classification: HYDROLASE Ligands: GOL |
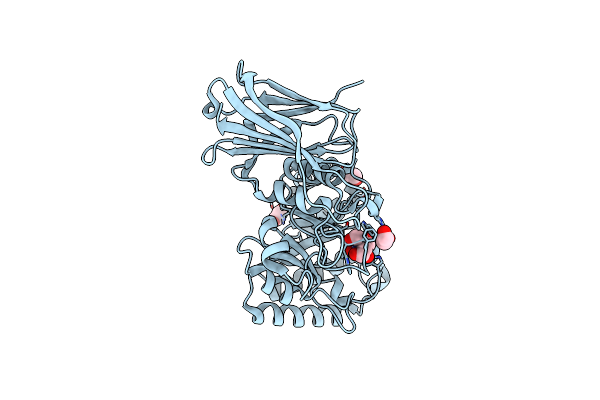 |
Organism: Neocallimastix frontalis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2017-06-14 Classification: HYDROLASE Ligands: BTB, EDO, IOD |
 |
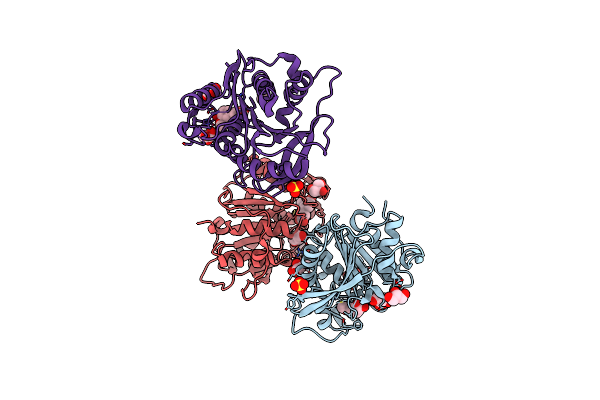
Structure Of The Ce1 Ferulic Acid Esterase Amce1/Fae1A, From The Anaerobic Fungi Anaeromyces Mucronatus In The Absence Of Substrate
Organism: Anaeromyces mucronatus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-04-27 Classification: HYDROLASE Ligands: GOL |
 |
Structure Of A Ce1 Ferulic Acid Esterase, Amce1/Fae1A, From Anaeromyces Mucronatus In Complex With Ferulic Acid
Organism: Anaeromyces mucronatus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2016-04-27 Classification: HYDROLASE Ligands: FER, SO4, GOL |
 |
Crystal Structure Of S. Aureus Tarm G117R Mutant In Complex With Udp And Udp-Glcnac
Organism: Staphylococcus aureus subsp. aureus 21178
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-03-04 Classification: TRANSFERASE Ligands: UDP, UD1 |
 |
Organism: Staphylococcus aureus subsp. aureus 21178
Method: X-RAY DIFFRACTION Resolution:3.19 Å Release Date: 2015-02-25 Classification: TRANSFERASE Ligands: UDP |
 |
Crystal Structure Of S. Aureus Tarm G117R Mutant In Complex With Fondaparinux, Alpha-Glcnac-Glycerol And Udp
Organism: Staphylococcus aureus subsp. aureus 21178
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2015-02-25 Classification: TRANSFERASE Ligands: UDP, 3YW |
 |
Organism: Staphylococcus aureus subsp. aureus 21178
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2015-02-18 Classification: TRANSFERASE Ligands: SO4 |
 |
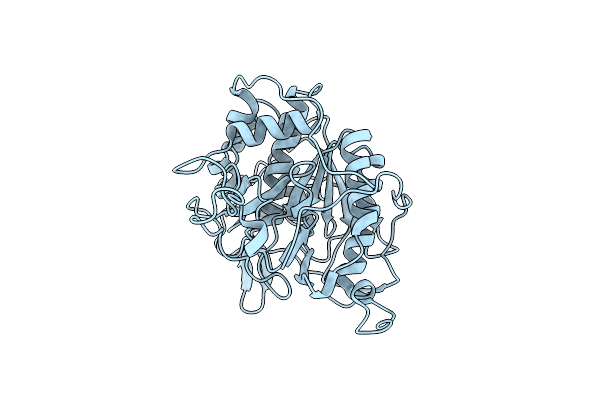
Organism: Bdellovibrio bacteriovorus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-07-23 Classification: HYDROLASE Ligands: MG, GOL |
 |
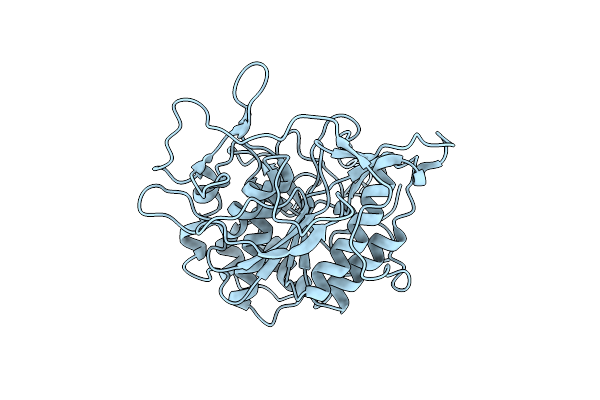
Organism: Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2013-05-22 Classification: HYDROLASE |
 |
Organism: Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2013-05-01 Classification: HYDROLASE |
 |
Structure Of The Ptp-Like Phytase From Selenomonas Ruminantium In Complex With Myo-Inositol (1,3,4,5)Tetrakisphosphate
Organism: Selenomonas ruminantium
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2011-12-07 Classification: HYDROLASE Ligands: ACT, GOL, 4IP, CL |
 |
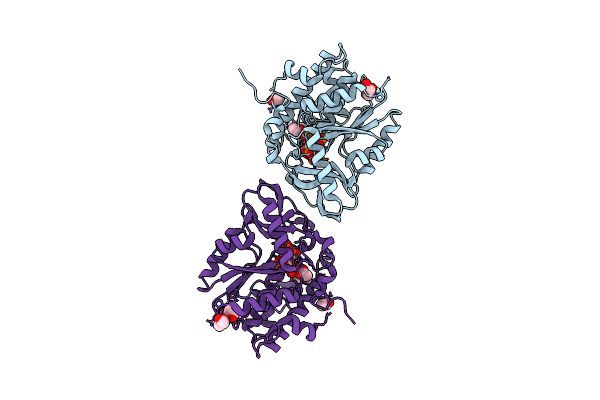
Structure Of The Ptp-Like Phytase From Selenomonas Ruminantium In Complex With Myo-Inositol (1,2,3,5,6)Pentakisphosphate
Organism: Selenomonas ruminantium
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2011-06-22 Classification: HYDROLASE Ligands: PO4, 5IP, GOL, ACT, CL |
 |
Structure Of The Ptp-Like Phytase From Selenomonas Ruminantium In Complex With Myo-Inositol Hexakisphosphate
Organism: Selenomonas ruminantium
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2011-06-15 Classification: HYDROLASE Ligands: IHP, PO4, ACT, GOL, CL |
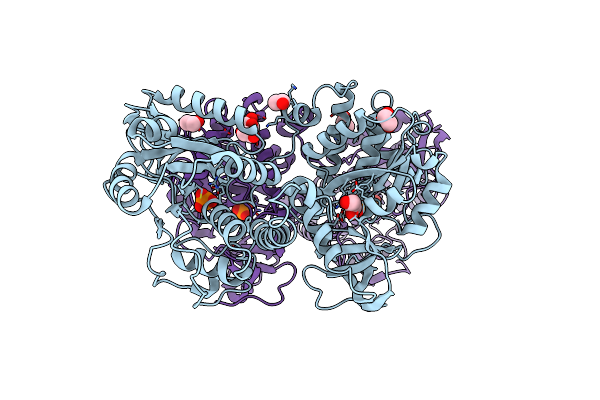 |
Structure Of The Tandemly Repeated Protein Tyrosine Phosphatase Like Phytase From Mitsuokella Multacida
Organism: Mitsuokella multacida
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-06-09 Classification: HYDROLASE Ligands: PO4, EDO |
 |
Structure Of The Ptp-Like Phytase Expressed By Selenomonas Ruminantium At An Ionic Strength Of 500 Mm
Organism: Selenomonas ruminantium
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-06-24 Classification: HYDROLASE Ligands: GOL |
 |
Structure Of The Ptp-Like Phytase Expressed By Selenomonas Ruminantium At An Ionic Strength Of 300 Mm
Organism: Selenomonas ruminantium
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-06-24 Classification: HYDROLASE Ligands: CL, GOL |
 |
Structure Of The Ptp-Like Phytase Expressed By Selenomonas Ruminantium At An Ionic Strength Of 400 Mm
Organism: Selenomonas ruminantium
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-06-24 Classification: HYDROLASE Ligands: CL, GOL |

