Search Count: 23
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2017-07-19 Classification: TRANSFERASE Ligands: CA |
 |
Crystal Structure Of Dapk1 Catalytic Domain In Complex With The Hinge Binding Fragment 4-Methylpyridazine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2015-05-13 Classification: TRANSFERASE Ligands: CL, SO4, DKG |
 |
Crystal Structure Of Dapk1 Catalytic Domain In Complex With The Hinge Binding Fragment Phthalazine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-05-06 Classification: TRANSFERASE Ligands: CL, ACT, SO4, 4FT |
 |
Crystal Structure Of P38 Alpha Map Kinase In Complex With A Novel Isoform Selective Drug Candidate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2015-02-25 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: GG5, 3GF, CL |
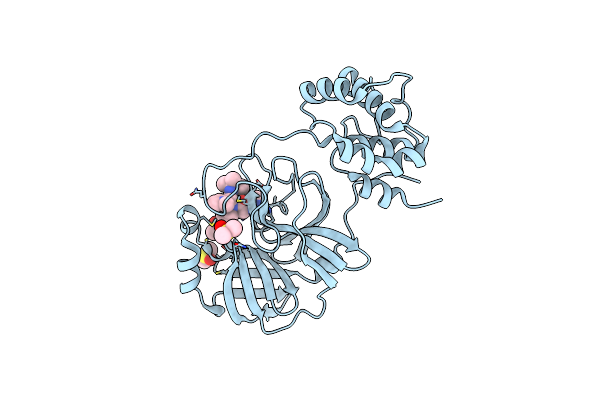 |
Discovery Of N-(Benzo[1,2,3]Triazol-1-Yl)-N-(Benzyl)Acetamido)Phenyl) Carboxamides As Severe Acute Respiratory Syndrome Coronavirus (Sars-Cov) 3Clpro Inhibitors: Identification Of Ml300 And Non-Covalent Nanomolar Inhibitors With An Induced-Fit Binding
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-10-02 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 23H, DMS |
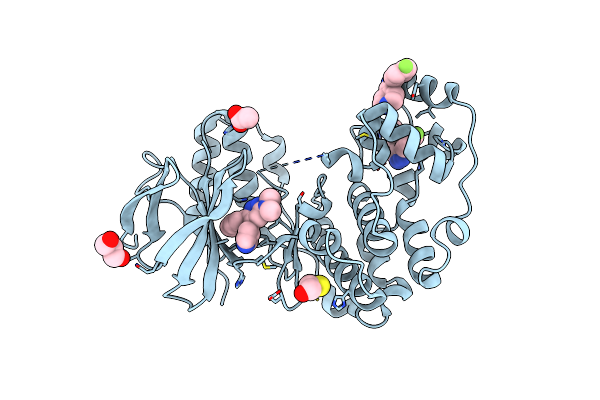 |
Human P38Alpha Mapk In Complex With A Novel And Selective Small Molecule Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-06-05 Classification: transferase/transferase inhibitor Ligands: GG5, LM4, ZN, ACT, BME, GOL |
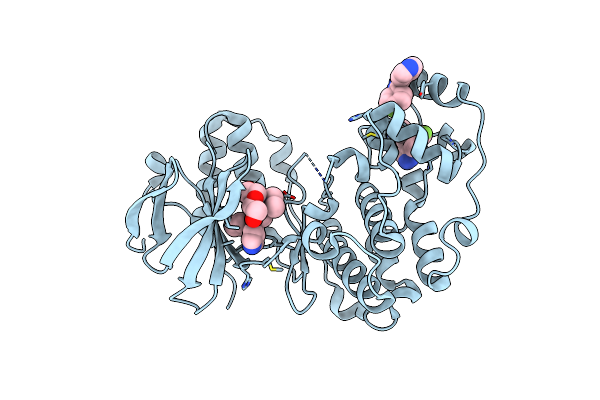 |
Human P38 Alpha Mapk In Complex With A Novel And Selective Small Molecule Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2013-06-05 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: GG5, LM3, BME, PEG |
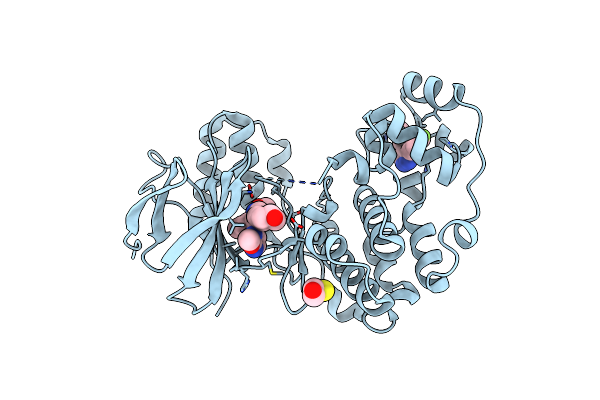 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-06-05 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: GG5, SB0, EDO, BME |
 |
Severe Acute Respiratory Syndrome Coronavirus (Sars-Cov) 3Cl Protease In Complex With N-[(1R)-2-(Tert-Butylamino)-2-Oxo-1-(Pyridin-3-Yl)Ethyl]-N-(4-Tert-Butylphenyl)Furan-2-Carboxamide Inhibitor.
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2013-01-16 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 0EN, DMS |
 |
Structure-Based Design And Synthesis And Biological Evaluation Of Peptidomimetic Sars-3Clpro Inhibitors
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-02-12 Classification: VIRAL PROTEIN Ligands: CYV |
 |
Crystal Structure Of A Junctionless All-Rna Hairpin Ribozyme At 2.05 Angstroms Resolution
Method: X-RAY DIFFRACTION
Resolution:2.05 Å Release Date: 2007-03-06 Classification: RNA Ligands: SO4, NCO |
 |
Crystal Structure Of A Novel Fructose 1,6-Bisphosphate And Alf3 Containing Pyrophosphate-Dependent Phosphofructo-1-Kinase Complex From Borrelia Burgdorferi
Organism: Borrelia burgdorferi
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2006-11-28 Classification: TRANSFERASE Ligands: FBP, AF3 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2006-08-29 Classification: HYDROLASE Ligands: CL, NA, GRL, GOL |
 |
The Crystal Structure Of An All-Rna Minimal Hairpin Ribozyme With Mutant G8I At The Cleavage Site
Method: X-RAY DIFFRACTION
Resolution:2.33 Å Release Date: 2006-02-14 Classification: RNA Ligands: SO4, NCO |
 |
The Structure Of An All-Rna Minimal Hairpin Ribozyme With Mutation G8A At The Cleavage Site
Method: X-RAY DIFFRACTION
Resolution:2.40 Å Release Date: 2006-02-14 Classification: RNA Ligands: SO4, NCO |
 |
The Structure Of A Minimal All-Rna Hairpin Ribozyme With The Mutant G8U At The Cleavage Site
Method: X-RAY DIFFRACTION
Resolution:2.38 Å Release Date: 2006-02-14 Classification: RNA Ligands: SO4, NCO |
 |
Crystal Structure Of A Minimal, Mutant All-Rna Hairpin Ribozyme (U39C, G8Mtu)
Method: X-RAY DIFFRACTION
Resolution:2.70 Å Release Date: 2006-02-14 Classification: RNA Ligands: NCO |
 |
Crystal Structure Of A Minimal, Mutant All-Rna Hairpin Ribozyme (U39C, G8I, 2'Deoxy A-1)
Method: X-RAY DIFFRACTION
Resolution:2.40 Å Release Date: 2006-02-14 Classification: RNA Ligands: SO4, NCO |
 |
Crystal Structure Of A Minimal, All Rna Hairpin Ribozyme With Modifications (G8Dap, U39C) At Ph 8.6
Method: X-RAY DIFFRACTION
Resolution:2.40 Å Release Date: 2006-02-14 Classification: RNA Ligands: NCO |
 |
Method: X-RAY DIFFRACTION
Resolution:2.19 Å Release Date: 2005-11-22 Classification: RNA Ligands: SO4, NCO |

