Search Count: 18
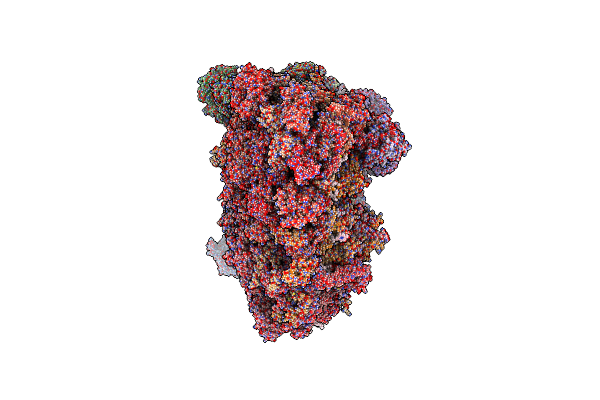 |
Cryo-Em Structure Of The I-Ii-Iii2-Iv2 Respiratory Supercomplex From Tetrahymena Thermophila
|
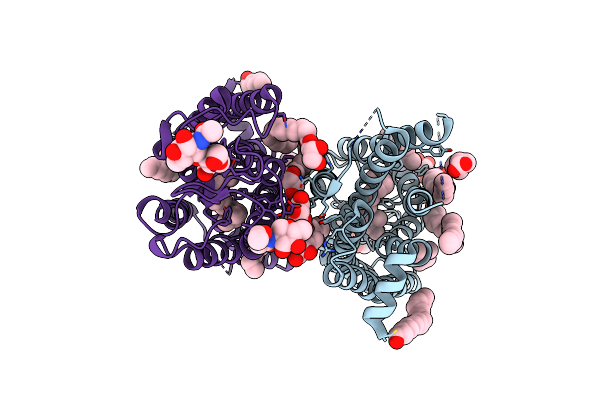 |
Dark State Crystal Structure Of Bovine Rhodopsin In Lipidic Cubic Phase (Sacla)
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-03-29 Classification: MEMBRANE PROTEIN Ligands: ACE, RET, NAG, DAO, OLC, PLM |
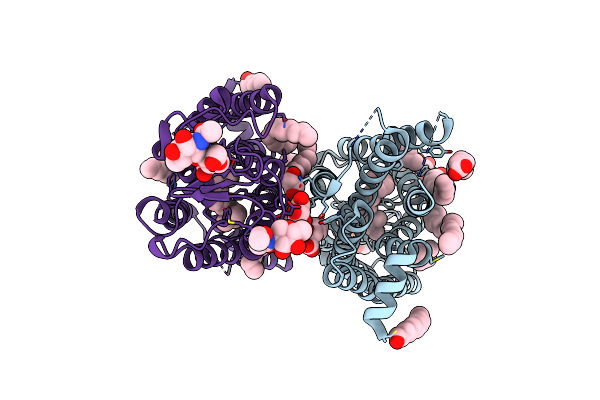 |
Dark State Crystal Structure Of Bovine Rhodopsin In Lipidic Cubic Phase (Swissfel)
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-03-29 Classification: MEMBRANE PROTEIN Ligands: ACE, RET, NAG, DAO, OLC, PLM |
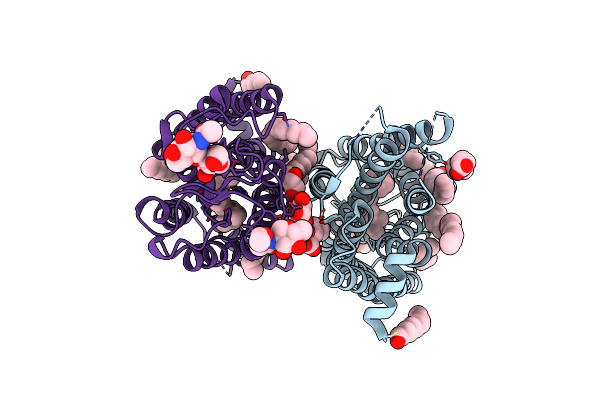 |
1 Picosecond Light Activated Crystal Structure Of Bovine Rhodopsin In Lipidic Cubic Phase
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-03-29 Classification: MEMBRANE PROTEIN Ligands: ACE, RET, NAG, DAO, OLC, PLM |
 |
10 Picosecond Light Activated Crystal Structure Of Bovine Rhodopsin In Lipidic Cubic Phase
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-03-29 Classification: MEMBRANE PROTEIN Ligands: ACE, RET, NAG, DAO, OLC, PLM |
 |
100 Picosecond Light Activated Crystal Structure Of Bovine Rhodopsin In Lipidic Cubic Phase (Sacla)
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-03-29 Classification: MEMBRANE PROTEIN Ligands: RET, NAG, DAO, OLC, PLM |
 |
Cryo-Em Structure Of Nadh:Ubiquinone Oxidoreductase (Complex-I) From Respiratory Supercomplex Of Tetrahymena Thermophila
Organism: Tetrahymena thermophila sb210
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: ELECTRON TRANSPORT Ligands: CDL, UDP, MG, PC1, NDP, 3PE, LPP, ADP, FES, SF4, FMN, 8Q1, ZN |
 |
Cryo-Em Structure Of Succinate Dehydrogenase Complex (Complex-Ii) In Respiratory Supercomplex Of Tetrahymena Thermophila
Organism: Tetrahymena thermophila
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: ELECTRON TRANSPORT Ligands: CDL, CA, FAD, FES, SF4, F3S, 3PE, HEC, PC1, UQ8 |
 |
Cryo-Em Structure Of Cytochrome C Oxidase Dimer (Complex Iv) From Respiratory Supercomplex Of Tetrahymena Thermophila
Organism: Tetrahymena thermophila sb210
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: ELECTRON TRANSPORT Ligands: HEA, CU, MG, CDL, PC1, 3PE, LPP, CA, CUA, ZN, K, UQ8, HEM, FES, ATP |
 |
Cryo-Em Structure Of Cytochrome Bc1 Complex (Complex-Iii) From Respiratory Supercomplex Of Tetrahymena Thermophila
Organism: Tetrahymena thermophila sb210
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: ELECTRON TRANSPORT Ligands: PC1, HEM, CDL, UQ8, PEE, HEC, FES |
 |
Femtosecond To Millisecond Structural Changes In A Light-Driven Sodium Pump: 20Ms Structure Of Kr2 With Extrapolated, Light And Dark Datasets
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA, NA |
 |
Femtosecond To Millisecond Structural Changes In A Light-Driven Sodium Pump: 1Ms Structure Of Kr2 With Extrapolated, Light And Dark Datasets
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA, NA |
 |
Femtosecond To Millisecond Structural Changes In A Light-Driven Sodium Pump: 30Us+150Us Structure Of Kr2 With Extrapolated, Light And Dark Datasets
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA |
 |
Femtosecond To Millisecond Structural Changes In A Light-Driven Sodium Pump: 1Ns+16Ns Structure Of Kr2 With Extrapolated, Light And Dark Datasets
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA |
 |
Femtosecond To Millisecond Structural Changes In A Light-Driven Sodium Pump: 800Fs+2Ps Structure Of Kr2 With Extrapolated, Light And Dark Datasets
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA |
 |
Femtosecond To Millisecond Structural Changes In A Light-Driven Sodium Pump: Dark Structure In Neutral Conditions With Attached Light Datasets At 800Fs, 2Ps, 100Ps, 1Ns, 16Ns, 1Us, 30Us, 150Us, 1Ms And 20Ms
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA |
 |
Femtosecond To Millisecond Structural Changes In A Light-Driven Sodium Pump: Dark Structure In Acidic Conditions
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA |
 |
Organism: Bos taurus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.12 Å Release Date: 2018-10-03 Classification: SIGNALING PROTEIN Ligands: RET, NAG |

