Search Count: 22
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-04-29 Classification: ACTIN-BINDING PROTEIN Ligands: SO4, EDO, DHL |
 |
Crystal Structure Of Retro-Aldolase Ra110.4-6 Complexed With Inhibitor 1-(6-Methoxy-2-Naphthalenyl)-1,3-Butanedione
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-11-12 Classification: ISOMERASE Ligands: LLK |
 |
Crystal Structure Of Kemp Eliminase Hg3.17 E47N,N300D Complexed With Transition State Analog 6-Nitrobenzotriazole
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2013-10-16 Classification: LYASE/LYASE INHIBITOR Ligands: SO4, 6NT |
 |
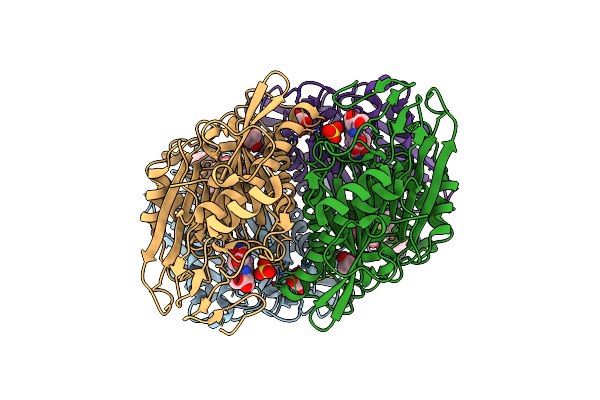
Crystal Structure Of The N-Terminal Beta-Aminopeptidase Bapa In Complex With Ampicillin
Organism: Sphingosinicella xenopeptidilytica
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-09-28 Classification: HYDROLASE/ANTIBIOTIC Ligands: GOL, AIC, NDV |
 |
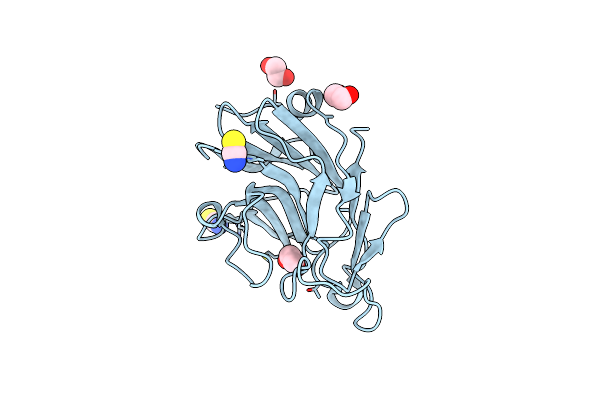
Crystal Structure Of The N-Terminal Beta-Aminopeptidase Bapa In Complex With Hydrolyzed Ampicillin
Organism: Sphingosinicella xenopeptidilytica
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2011-09-28 Classification: Hydrolase/Inhibitor Ligands: GOL, OAE, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2009-10-20 Classification: SIGNALING PROTEIN Ligands: SCN, EDO |
 |
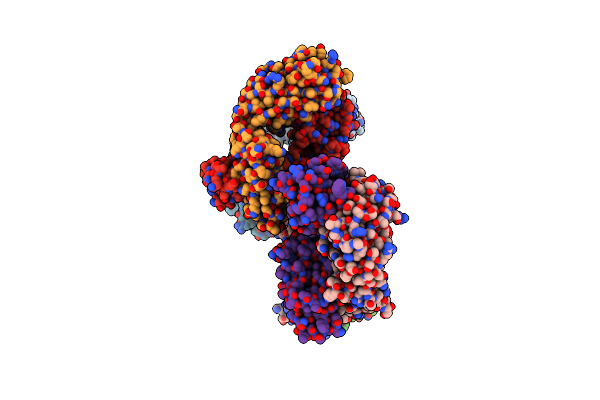
The Missing Part Of The Bacterial Mexab-Oprm System: Structural Determination Of The Multidrug Exporter Mexb
Organism: Pseudomonas aeruginosa pa01
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2009-05-19 Classification: MEMBRANE PROTEIN Ligands: LMT |
 |
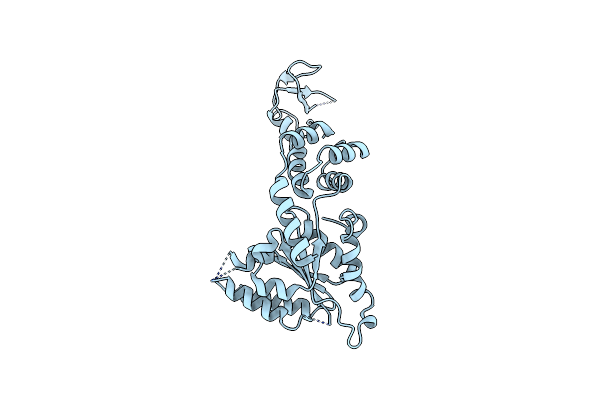
Unusual Twinning In Crystals Of The Cits Binding Antibody Fab Fragment F3P4
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2008-06-17 Classification: IMMUNE SYSTEM |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2008-02-26 Classification: PROTEIN TRANSPORT |
 |
Crystal Structure Of An N-Terminal Deletion Mutant Of Escherichia Coli Gadb In An Autoinhibited State (Aldamine)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2006-06-20 Classification: LYASE Ligands: SO4, PLP, EDO |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2006-06-20 Classification: LYASE Ligands: BR, PLP, ACY |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2006-06-20 Classification: LYASE Ligands: IOD, PLP, FMT, ACY, PEG |
 |
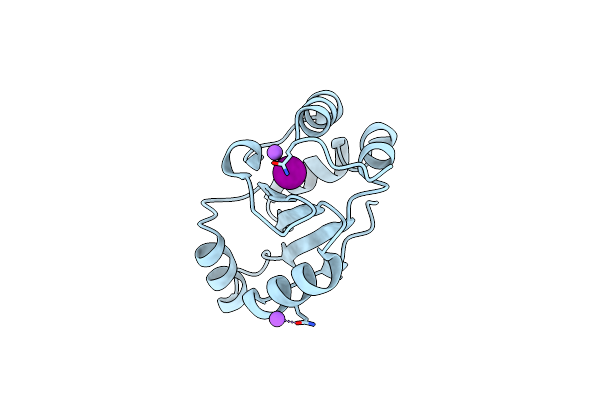
Crystal Structure Of The C-Terminal Domain Of The Electron Transfer Catalyst Dsbd (Oxidized Form)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2006-06-13 Classification: OXIDOREDUCTASE Ligands: IOD, NI, NA |
 |
High Resolution Crystal Structure Of The C-Terminal Domain Of The Electron Transfer Catalyst Dsbd (Reduced Form)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2006-06-13 Classification: OXIDOREDUCTASE Ligands: IOD, NA |
 |
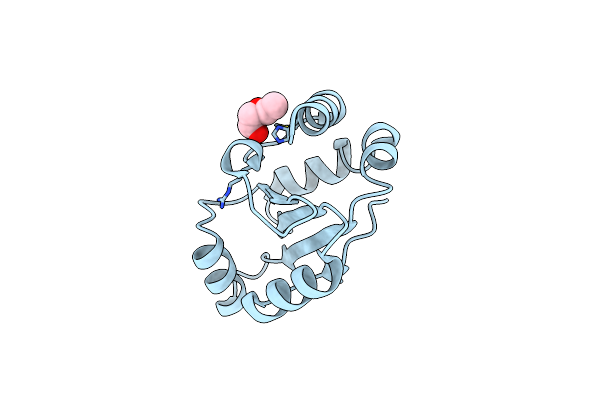
High Resolution Crystal Structure Of The C-Terminal Domain Of The Electron Transfer Catalyst Dsbd (Photoreduced Form)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2006-06-13 Classification: OXIDOREDUCTASE |
 |
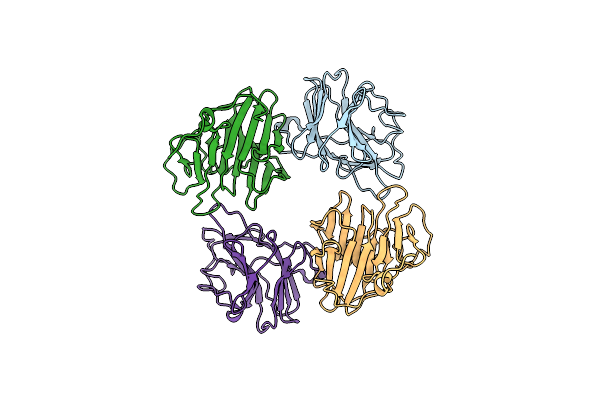
Atomic Resolution Crystal Structure Of The C-Terminal Domain Of The Electron Transfer Catalyst Dsbd (Reduced Form At Ph7)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:0.99 Å Release Date: 2006-06-13 Classification: OXIDOREDUCTASE Ligands: IOD, PEG |
 |
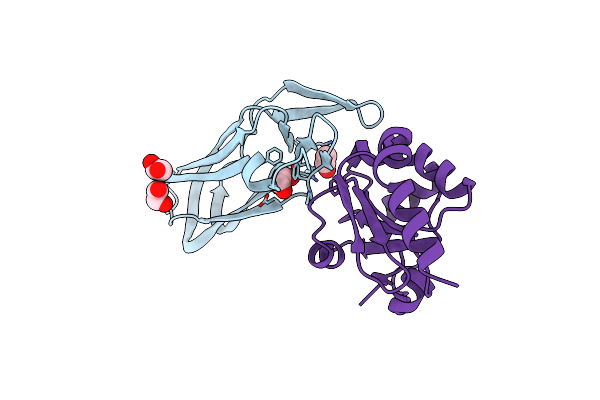
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2006-01-10 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of The Disulfide-Linked Complex Between The N-Terminal Domain Of The Electron Transfer Catalyst Dsbd And The Cytochrome C Biogenesis Protein Ccmg
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2005-07-19 Classification: OXIDOREDUCTASE/BIOSYNTHETIC PROTEIN Ligands: CL, EDO |
 |
Crystal Structure Of The Disulfide-Linked Complex Between The N-Terminal And C-Terminal Domain Of The Electron Transfer Catalyst Dsbd
Organism: Escherichia coli str. k12 substr.
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2005-07-12 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of A Designed Selected Ankyrin Repeat Protein In Complex With The Maltose Binding Protein
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2004-05-25 Classification: de novo protein/sugar binding protein |

