Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2020-10-14 Classification: OXIDOREDUCTASE Ligands: HEM, RWZ, GOL, TRS, MG, FMT |
 |
Fragment Of Nitrate/Nitrite Sensor Histidine Kinase Narq (Wt) In Symmetric Holo State
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2017-05-31 Classification: TRANSFERASE Ligands: LFA, NO3 |
 |
Fragment Of Nitrate/Nitrite Sensor Histidine Kinase Narq (Wt) In Asymmetric Holo State
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2017-05-31 Classification: TRANSFERASE Ligands: NO3, LFA |
 |
Fragment Of Nitrate/Nitrite Sensor Histidine Kinase Narq (R50K) In Symmetric Apo State
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-05-31 Classification: TRANSFERASE Ligands: PO4 |
 |
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-02-15 Classification: SIGNALING PROTEIN Ligands: RET, BOG, LFA |
 |
Structure Of The Srii/Htrii Complex In I212121 Space Group ("U" Shape) - M State
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-02-15 Classification: SIGNALING PROTEIN Ligands: RET, BOG, LFA |
 |
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-02-15 Classification: SIGNALING PROTEIN Ligands: RET, LFA |
 |
Structure Of The Srii/Htrii(G83F) Complex In P212121 Space Group ("V" Shape)
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2017-02-15 Classification: SIGNALING PROTEIN Ligands: LFA, BOG, RET |
 |
Crystal Structure Of Archaeoglobus Fulgidus Ipct-Dipps Bifunctional Membrane Protein
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2014-07-02 Classification: TRANSFERASE Ligands: LFA, MG |
 |
Structure Of The Srii(D75N Mutant)/Htrii Complex In I212121 Space Group ("U" Shape)
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2013-05-08 Classification: MEMBRANE PROTEIN Ligands: RET, LFA |
 |
Crystal Structure Of The Complex Of The Carbohydrate Recognition Domain Of Human Dc-Sign With Pseudo Trimannoside Mimic.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2011-10-19 Classification: SUGAR BINDING PROTEIN Ligands: MAN, 07B, AE9, CA, CL |
 |
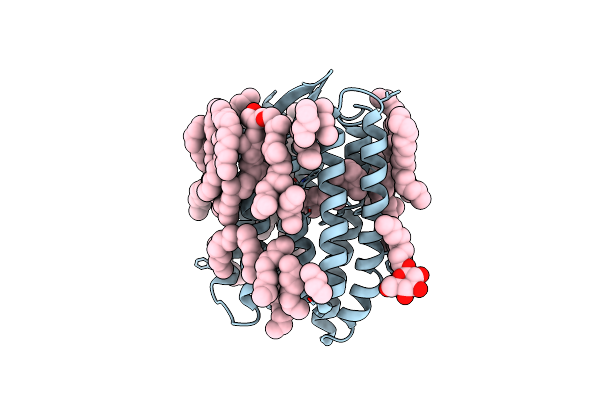
Crystal Structure Of Natronomonas Pharaonis Sensory Rhodopsin Ii In The Ground State
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-09-14 Classification: TRANSPORT PROTEIN Ligands: RET, BOG, LFA, L2P |
 |
Crystal Structure Of Natronomonas Pharaonis Sensory Rhodopsin Ii In The Active State
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-09-14 Classification: MEMBRANE PROTEIN Ligands: RET, BOG, LFA, L2P |