Search Count: 12
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-06-23 Classification: RIBOSOME Ligands: AGS, TDB |
 |
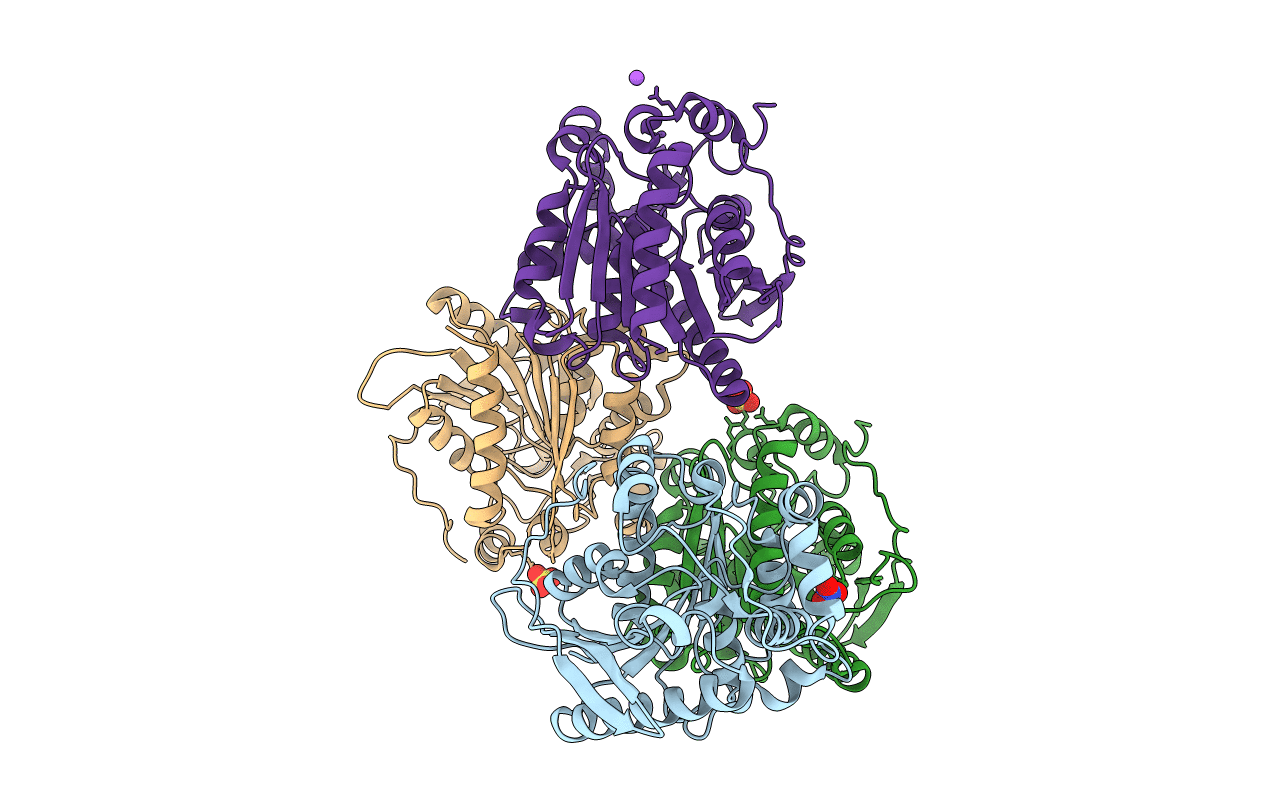
Crystal Structure Of A Soluble Variant Of Monoglyceride Lipase From Saccharomyces Cerevisiae In Complex With A Substrate Analog
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-05-25 Classification: HYDROLASE Ligands: SO4, 4S7, NO3 |
 |
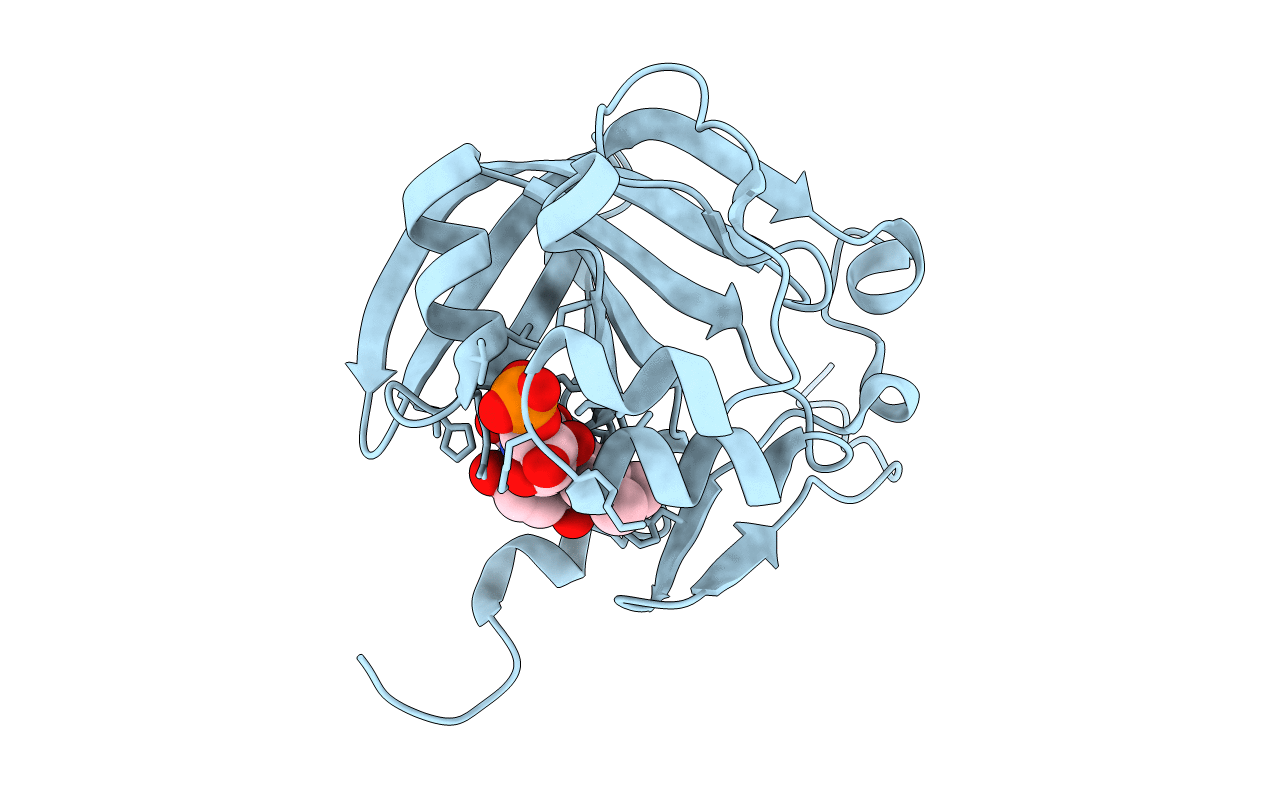
Crystal Structure Of A Soluble Variant Of The Monoglyceride Lipase From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2016-04-27 Classification: HYDROLASE Ligands: NO3, SO4, NA |
 |
Crystal Structure Of Fmn-Binding Protein (Np_142786.1) From Pyrococcus Horikoshii With Bound P-Hydroxybenzaldehyde
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-05-14 Classification: FMN-BINDING PROTEIN Ligands: FMN, HBA |
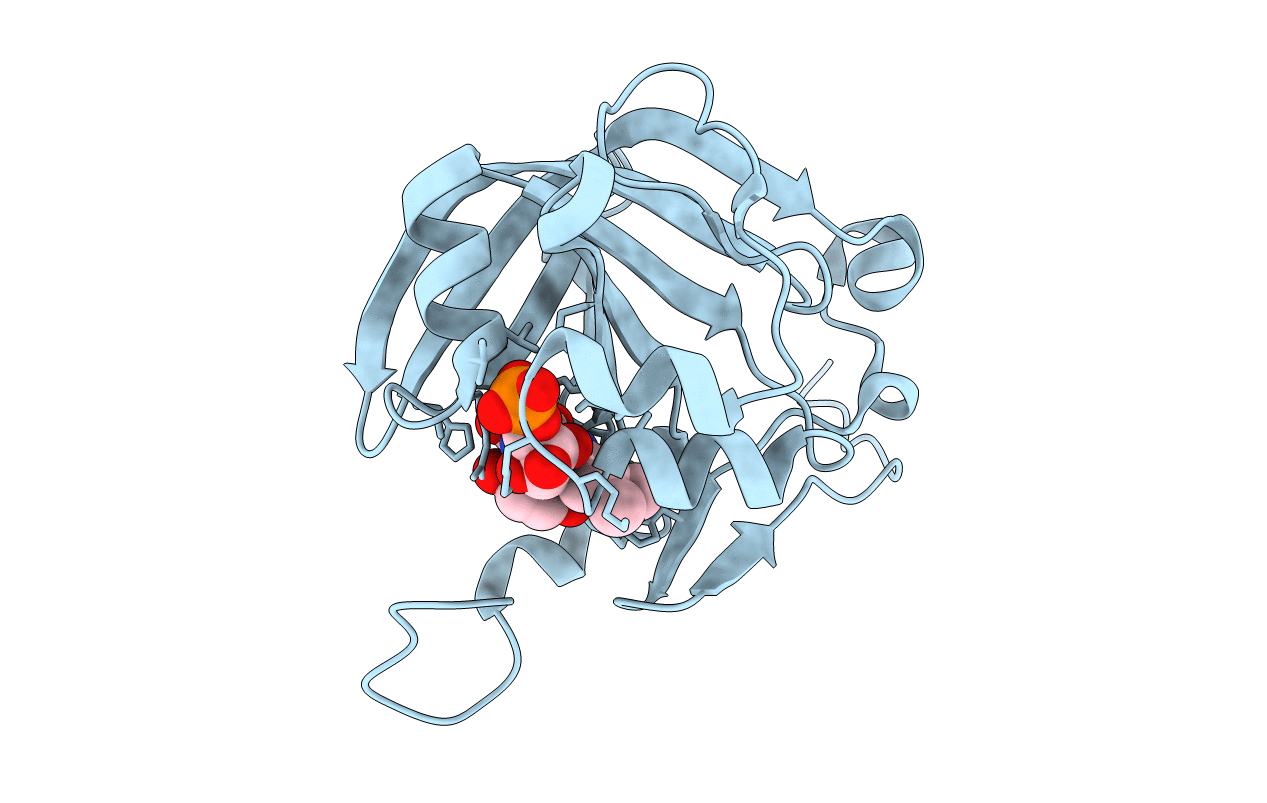 |
Crystal Structure Of Fmn-Binding Protein (Np_142786.1) From Pyrococcus Horikoshii With Bound Benzene-1,4-Diol
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2014-05-14 Classification: FMN-BINDING PROTEIN Ligands: FMN, HQE |
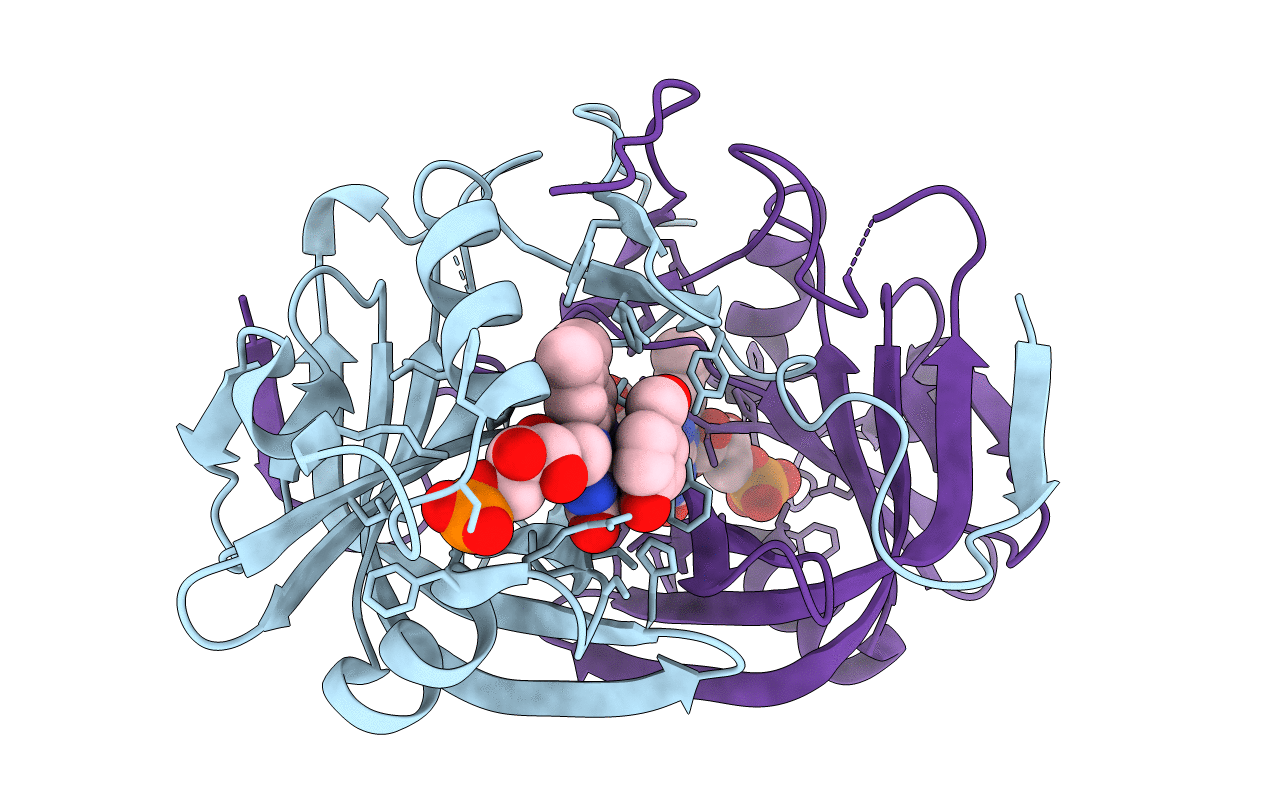 |
Crystal Structure Of Fmn-Binding Protein (Yp_005476) From Thermus Thermophilus With Bound P-Hydroxybenzaldehyde
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2014-05-14 Classification: FMN-BINDING PROTEIN Ligands: HBA, FMN |
 |
Crystal Structure Of Fmn-Binding Protein (Yp_005476) From Thermus Thermophilus With Bound Benzene-1,4-Diol
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-05-14 Classification: FMN-BINDING PROTEIN Ligands: HQE, FMN |
 |
Crystal Structure Of Fmn-Binding Protein (Np_142786.1) From Pyrococcus Horikoshii With Bound 1-Cyclohex-2-Enone
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2014-05-14 Classification: OXIDOREDUCTASE Ligands: FMN, A2Q |
 |
Crystal Structure Of Fmn-Binding Protein (Yp_005476) From Thermus Thermophilus With Bound 1-Cyclohex-2-Enone
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-05-14 Classification: FMN-BINDING PROTEIN Ligands: A2Q, FMN |
 |
Ntf2-Like, Potential Transfer Protein Tram From Gram-Positive Conjugative Plasmid Pip501
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-12-05 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Monoacylglycerol Lipase From Bacillus Sp. H257 In Complex With Pmsf
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2012-05-23 Classification: HYDROLASE Ligands: MRD, PMS |
 |
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2012-05-02 Classification: HYDROLASE Ligands: MRD |

