Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
M. Tuberculosis Meets European Lead Factory: Identification And Structural Characterization Of Novel Rv0183 Inhibitors Using X-Ray Crystallography: Elf5
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: HYDROLASE Ligands: 8KE, I3F, CL |
 |
M. Tuberculosis Meets European Lead Factory: Identification And Structural Characterization Of Novel Rv0183 Inhibitors Using X-Ray Crystallography: Elf8
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: HYDROLASE Ligands: 8D5, MPD, EDO, PGE, PEG |
 |
M. Tuberculosis Meets European Lead Factory: Identification And Structural Characterization Of Novel Rv0183 Inhibitors Using X-Ray Crystallography: Elf1
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: HYDROLASE Ligands: 7WW, PO4, MPD, DMS, EOH, EDO |
 |
Cova2-15 Fragment Antigen Binding In Complex With Sars-Cov-2 6P-Mut7 S Protein
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Crystal Structure Of Sars-Cov-2 Receptor Binding Domain In Complex With Neutralizing Antibody Cova2-15
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.38 Å Release Date: 2024-12-04 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens, Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: MEMBRANE PROTEIN |
 |
Organism: Oldenlandia affinis
Method: SOLUTION NMR Release Date: 2021-10-20 Classification: PLANT PROTEIN |
 |
Organism: Oldenlandia affinis
Method: SOLUTION NMR Release Date: 2021-10-20 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Psychotria vellosiana
Method: SOLUTION NMR Release Date: 2021-08-04 Classification: IMMUNOSUPPRESSANT |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-06-23 Classification: RIBOSOME Ligands: AGS, TDB |
 |
Solution Structure Of The Fus/Tls Rna Recognition Motif In Complex With U1 Snrna Stem Loop Iii
|
 |
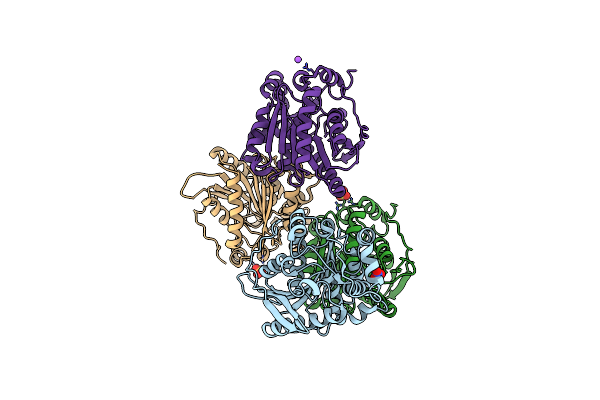
Crystal Structure Of A Soluble Variant Of Monoglyceride Lipase From Saccharomyces Cerevisiae In Complex With A Substrate Analog
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-05-25 Classification: HYDROLASE Ligands: SO4, 4S7, NO3 |
 |
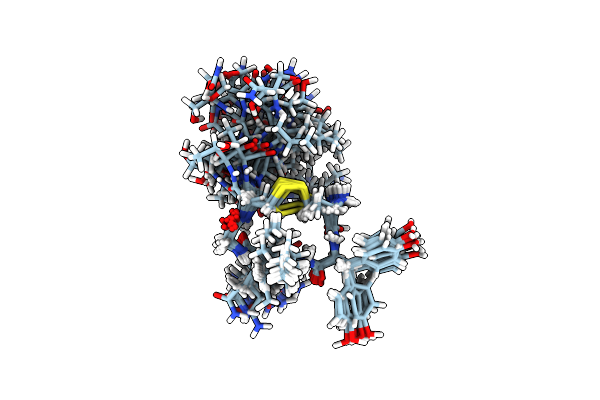
Crystal Structure Of A Soluble Variant Of The Monoglyceride Lipase From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2016-04-27 Classification: HYDROLASE Ligands: NO3, SO4, NA |
 |
 |
 |
Crystal Structure Of Fmn-Binding Protein (Np_142786.1) From Pyrococcus Horikoshii With Bound P-Hydroxybenzaldehyde
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-05-14 Classification: FMN-BINDING PROTEIN Ligands: FMN, HBA |
 |
Crystal Structure Of Fmn-Binding Protein (Np_142786.1) From Pyrococcus Horikoshii With Bound Benzene-1,4-Diol
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2014-05-14 Classification: FMN-BINDING PROTEIN Ligands: FMN, HQE |
 |
Crystal Structure Of Fmn-Binding Protein (Yp_005476) From Thermus Thermophilus With Bound P-Hydroxybenzaldehyde
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2014-05-14 Classification: FMN-BINDING PROTEIN Ligands: HBA, FMN |
 |
Crystal Structure Of Fmn-Binding Protein (Yp_005476) From Thermus Thermophilus With Bound Benzene-1,4-Diol
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-05-14 Classification: FMN-BINDING PROTEIN Ligands: HQE, FMN |
 |
Crystal Structure Of Fmn-Binding Protein (Np_142786.1) From Pyrococcus Horikoshii With Bound 1-Cyclohex-2-Enone
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2014-05-14 Classification: OXIDOREDUCTASE Ligands: FMN, A2Q |