Search Count: 16
 |
Metal-Binding Domain 3 Of Copper-Transporting Atpase Ran1 From Arabidopsis Thaliana
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-01-29 Classification: METAL BINDING PROTEIN |
 |
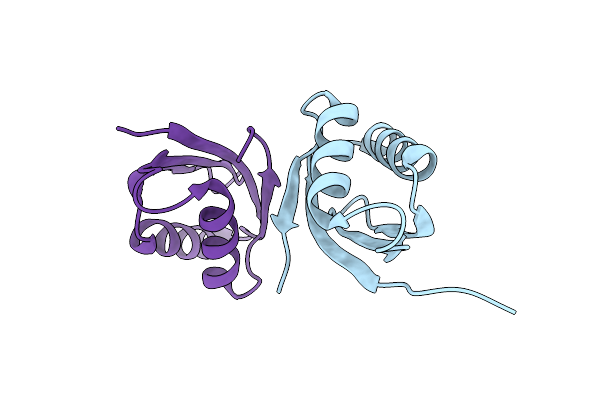
Crystal Structure Hr1 Domain Of Rho-Associated Coiled-Coil Protein Kinases (Rock-Hr1)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-06-21 Classification: SIGNALING PROTEIN Ligands: DTT |
 |
Mutant Nitrobindin M75L/H76L/Q96C/M148L (Nb4H) From Arabidopsis Thaliana With Cofactor Mnppix
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2021-05-26 Classification: TRANSPORT PROTEIN Ligands: EDO, MNH |
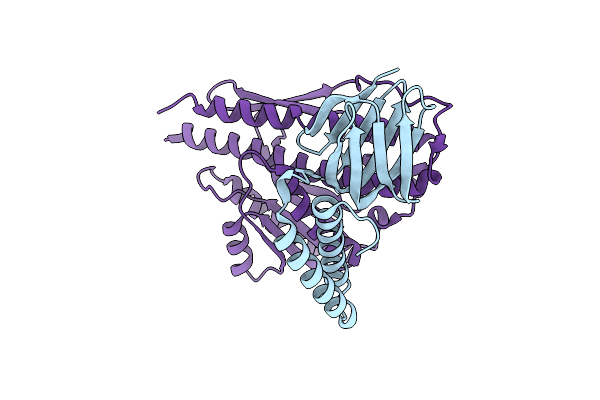 |
Crystal Structure Of The Gamma - Epsilon Complex Of Photosynthetic Cyanobacterial F1-Atpase
Organism: Thermosynechococcus elongatus (strain bp-1)
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2018-09-26 Classification: HYDROLASE |
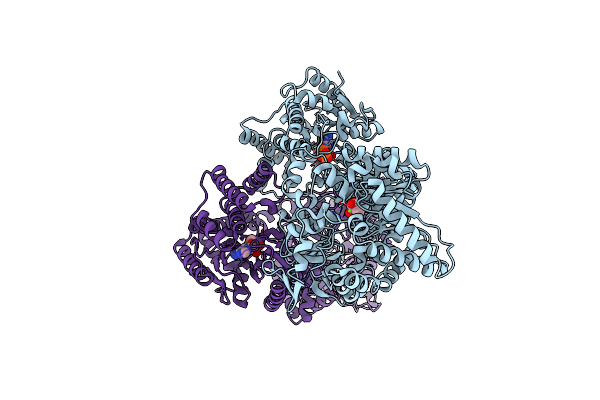 |
C4-Type Pyruvate Phosphate Dikinase: Conformational Intermediate Of Central Domain In The Swiveling Mechanism
Organism: Flaveria trinervia
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2017-05-24 Classification: TRANSFERASE Ligands: ADP, MG, PYR |
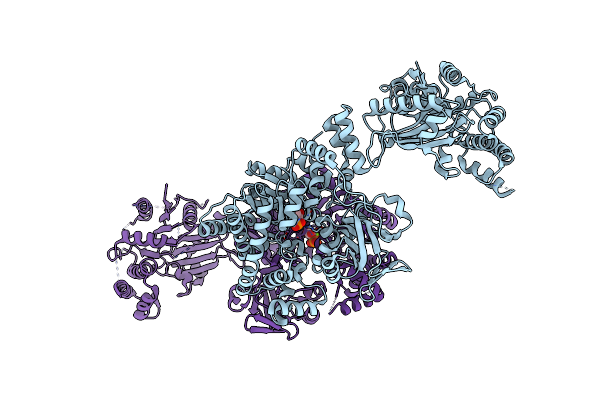 |
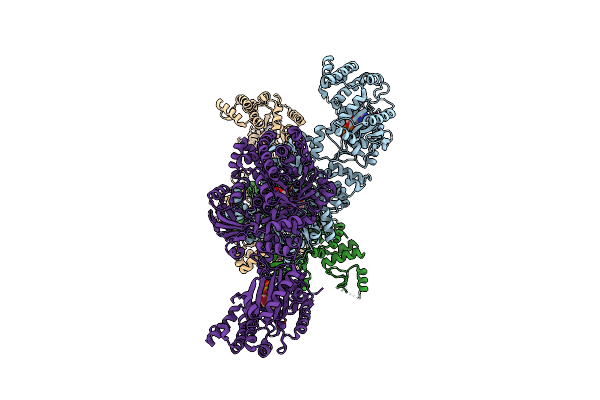
C4-Type Pyruvate Phosphate Dikinase: Different Conformational States Of The Nucleotide Binding Domain In The Dimer
Organism: Flaveria trinervia
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2017-04-05 Classification: TRANSFERASE Ligands: PEP, MG |
 |
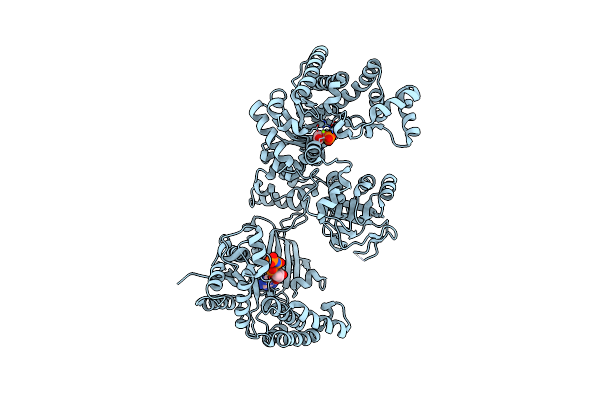
C4-Type Pyruvate Phospate Dikinase: Nucleotide Binding Domain With Bound Atp Analogue
Organism: Flaveria trinervia
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2017-04-05 Classification: TRANSFERASE Ligands: 6NQ, MG, PEP |
 |
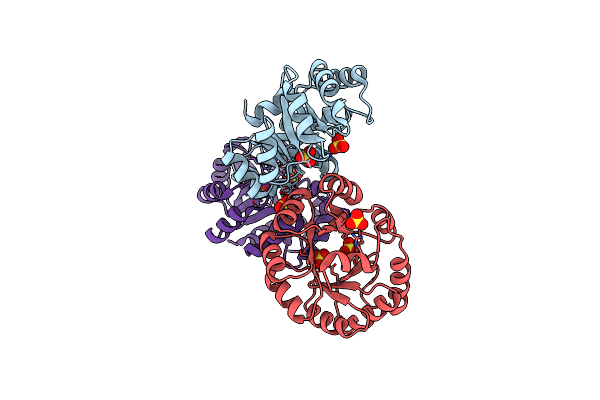
C3-Type Pyruvate Phosphate Dikinase: Intermediate State Of The Central Domain In The Swiveling Mechanism
Organism: Flaveria pringlei
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2017-04-05 Classification: TRANSFERASE Ligands: 6NQ, PEP, MG |
 |
Crystal Structure Of 2-Keto-3-Deoxy-6-Phospho-Gluconate Aldolase From Zymomonas Mobilis Atcc 29191
Organism: Zymomonas mobilis
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2014-04-30 Classification: LYASE Ligands: SO4 |
 |
Resolving The Activation Site Of Positive Regulators In Plant Phosphoenolpyruvate Carboxylase
Organism: Flaveria trinervia
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2013-10-02 Classification: LYASE Ligands: G6P, SO4, EDO, TRS |
 |
Resolving The Activation Site Of Positive Regulators In Plant Phosphoenolpyruvate Carboxylase
Organism: Flaveria trinervia
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2013-10-02 Classification: LYASE Ligands: EDO, SO4 |
 |
Greater Efficiency Of Photosynthetic Carbon Fixation Due To Single Amino Acid Substitution
Organism: Flaveria pringlei
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2013-02-27 Classification: LYASE Ligands: ASP, EDO, SO4 |
 |
Greater Efficiency Of Photosynthetic Carbon Fixation Due To Single Amino Acid Substitution
Organism: Flaveria trinervia
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2013-02-27 Classification: LYASE Ligands: ASP, SO4, EDO |
 |
Structure Of The C14-Rotor Ring Of The Proton Translocating Chloroplast Atp Synthase
Organism: Spinacia oleracea
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2009-05-19 Classification: HYDROLASE |
 |
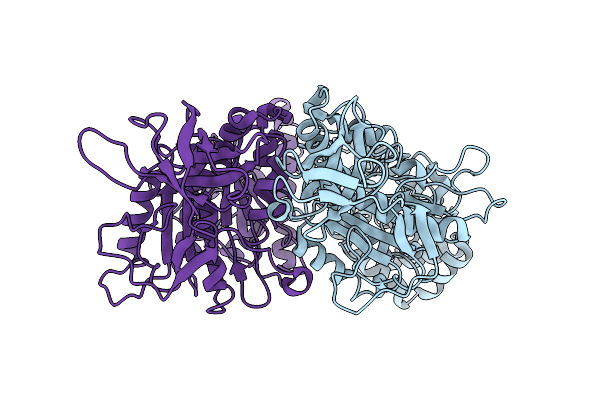
Organism: Spinacia oleracea
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2002-03-13 Classification: HYDROLASE Ligands: TTX |
 |
Organism: Spinacia oleracea
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2001-09-25 Classification: HYDROLASE |

