Search Count: 13
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2016-11-02 Classification: HYDROLASE Ligands: CA, BCN, NA |
 |
Crystal Structure Of B. Subtilis 168 Glpq In Complex With Glycerol-3-Phosphate (5 Minute Soak)
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2016-11-02 Classification: HYDROLASE Ligands: CA, G3P, NA |
 |
Crystal Structure Of B. Subtilis 168 Glpq In Complex With Glycerol-3-Phosphate (1 Hour Soak)
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2016-11-02 Classification: HYDROLASE Ligands: CA, G3P, PO4, NA |
 |
Structure Of The Human Quinone Reductase 2 (Nqo2) In Complex With Imiquimod
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2016-07-27 Classification: OXIDOREDUCTASE Ligands: ZN, FAD, 6T0, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2016-07-27 Classification: OXIDOREDUCTASE Ligands: ZN, FAD, SO4, C09 |
 |
Organism: Vibrio parahaemolyticus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-09-26 Classification: SIGNALING PROTEIN |
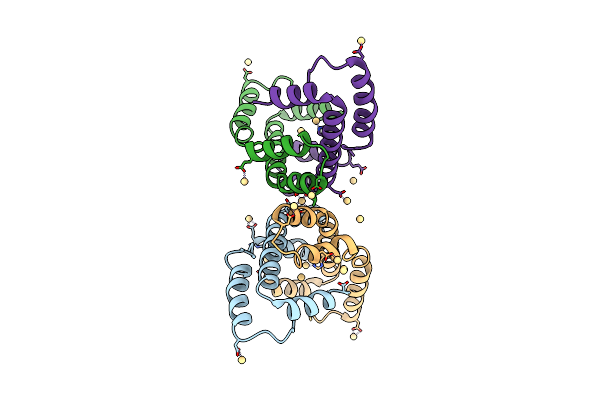 |
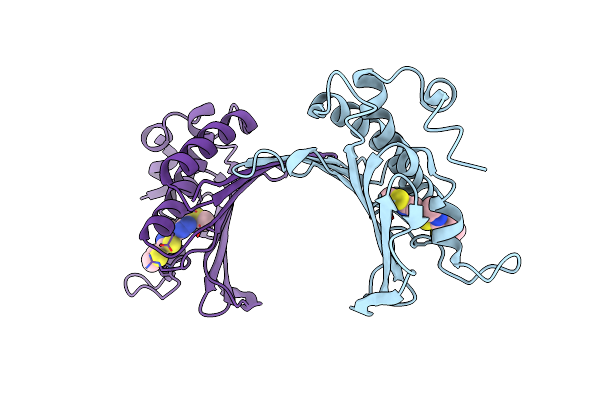
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2010-09-22 Classification: SIGNALING PROTEIN Ligands: CD |
 |
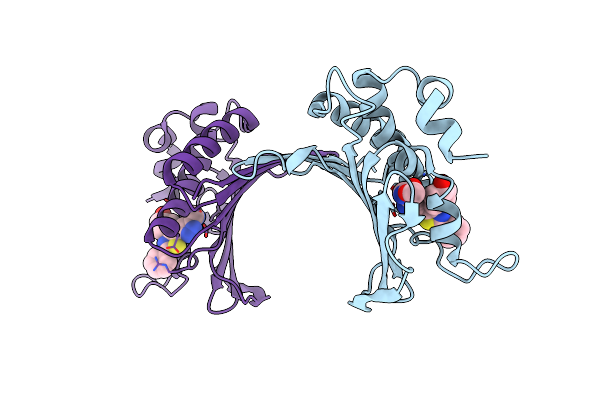
Crystal Structure Of Staphylococcus Aureus Gyrase B Co-Complexed With 4-Methyl-5-[3-(Methylsulfanyl)-1H-Pyrazol-5-Yl]-2-Thiophen-2-Yl-1,3-Thiazole Inhibitor
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-02-09 Classification: Isomerase/Isomerase inhibitor Ligands: B48 |
 |
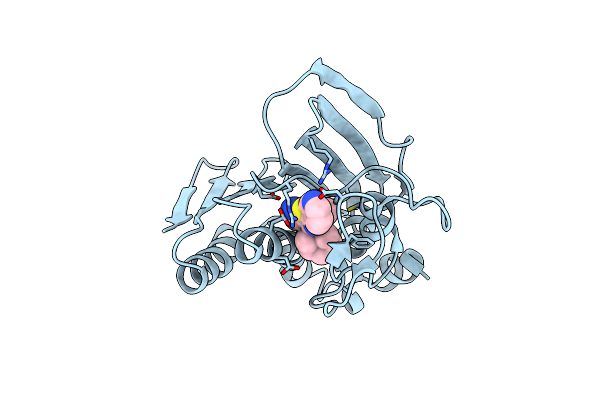
Staphylococcus Aureus Gyrase B Co-Complex With Methyl ({5-[4-(4-Hydroxypiperidin-1-Yl)-2-Phenyl-1,3-Thiazol-5-Yl]-1H-Pyrazol-3-Yl}Methyl)Carbamate Inhibitor
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-02-09 Classification: Isomerase/Isomerase inhibitor Ligands: B47 |
 |
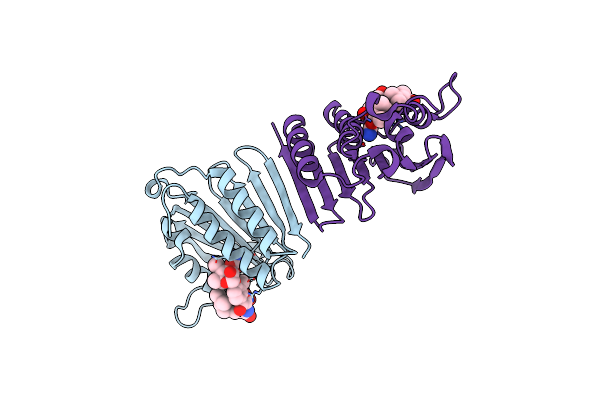
Crystal Structure Of E. Coli Gyrase B Co-Complexed With Prop-2-Yn-1-Yl {[5-(4-Piperidin-1-Yl-2-Pyridin-3-Yl-1,3-Thiazol-5-Yl)-1H-Pyrazol-3-Yl]Methyl}Carbamate Inhibitor
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-02-09 Classification: Isomerase/Isomerase inhibitor Ligands: B46 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-05-04 Classification: ISOMERASE Ligands: NOV |
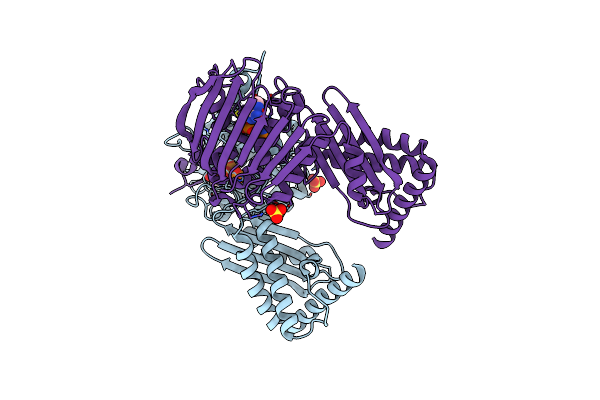 |
Crystal Structure Of E. Coli Topoisomerase Iv Pare 43Kda Subunit Complexed With Adpnp
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2004-05-04 Classification: ISOMERASE Ligands: MG, SO4, ANP |
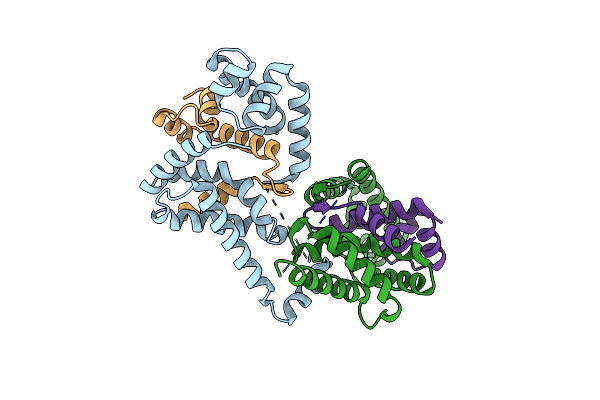 |
Crystal Structure Of Escherichia Coli Sigmae With The Cytoplasmic Domain Of Its Anti-Sigma Rsea
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-04-15 Classification: TRANSCRIPTION |

