Search Count: 18
 |
Sgbp-B From A Complex Xyloglucan Utilization Locus In Bacteroides Uniformis
Organism: Bacteroides uniformis (strain atcc 8492 / dsm 6597 / cip 103695 / jcm 5828 / nctc 13054 / vpi 0061)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-09-29 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Organism: Bacteroides uniformis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-09-13 Classification: HYDROLASE Ligands: EDO, TRS |
 |
Organism: Bacteroides uniformis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-09-13 Classification: HYDROLASE Ligands: GIF, EDO |
 |
Organism: Bacteroides uniformis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-09-13 Classification: HYDROLASE Ligands: EDO, SO4 |
 |
Organism: Bacteroides uniformis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-09-13 Classification: HYDROLASE Ligands: GIF, EDO |
 |
Organism: Bacteroides uniformis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-09-13 Classification: HYDROLASE |
 |
Organism: Bacteroides uniformis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2017-09-13 Classification: HYDROLASE Ligands: SO4, EDO, GOL, CA |
 |
Organism: Bacteroides uniformis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-09-13 Classification: HYDROLASE Ligands: CA, GOL, EDO |
 |
Organism: Bacteroides uniformis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2017-09-13 Classification: HYDROLASE Ligands: SO4, EDO, CA |
 |
Organism: Bacteroides uniformis
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2017-09-13 Classification: HYDROLASE Ligands: MG, EDO, TRS |
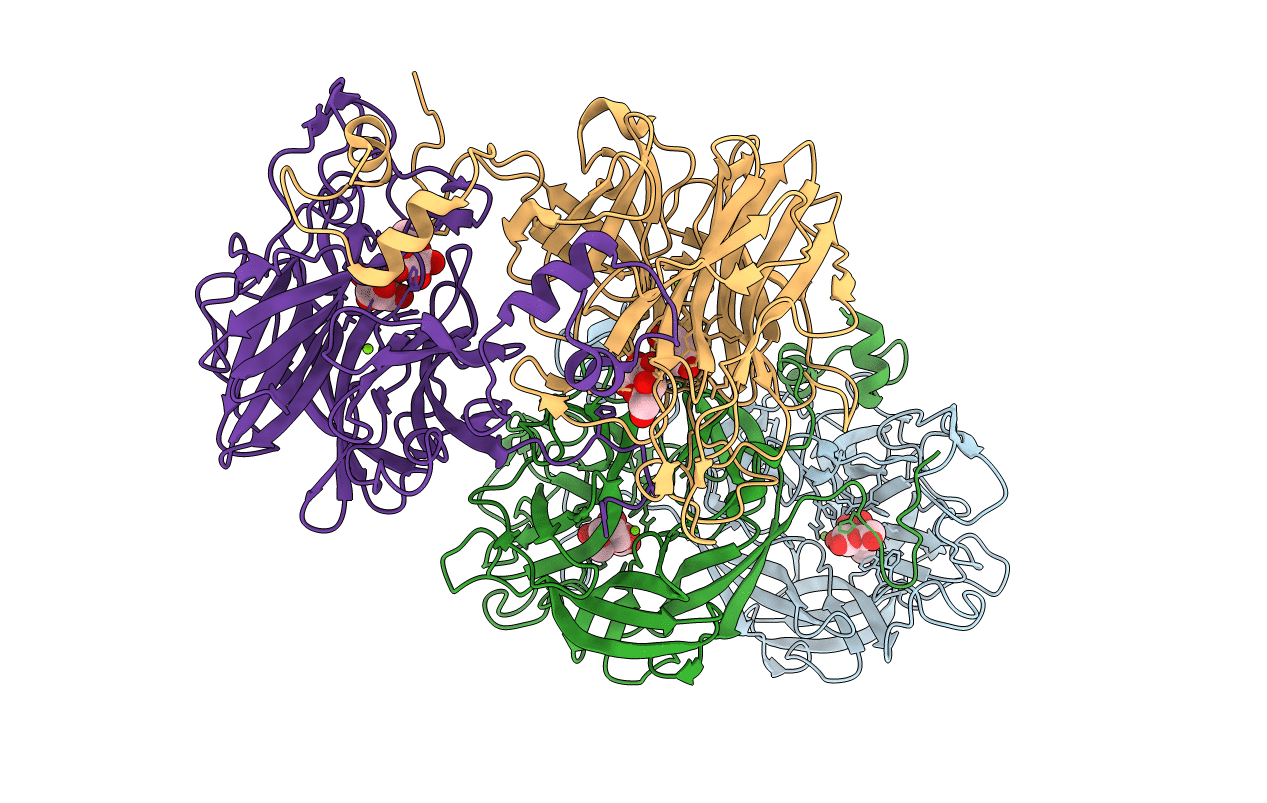 |
Organism: Bacteroides uniformis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-09-13 Classification: HYDROLASE Ligands: GAL, AAL, MG |
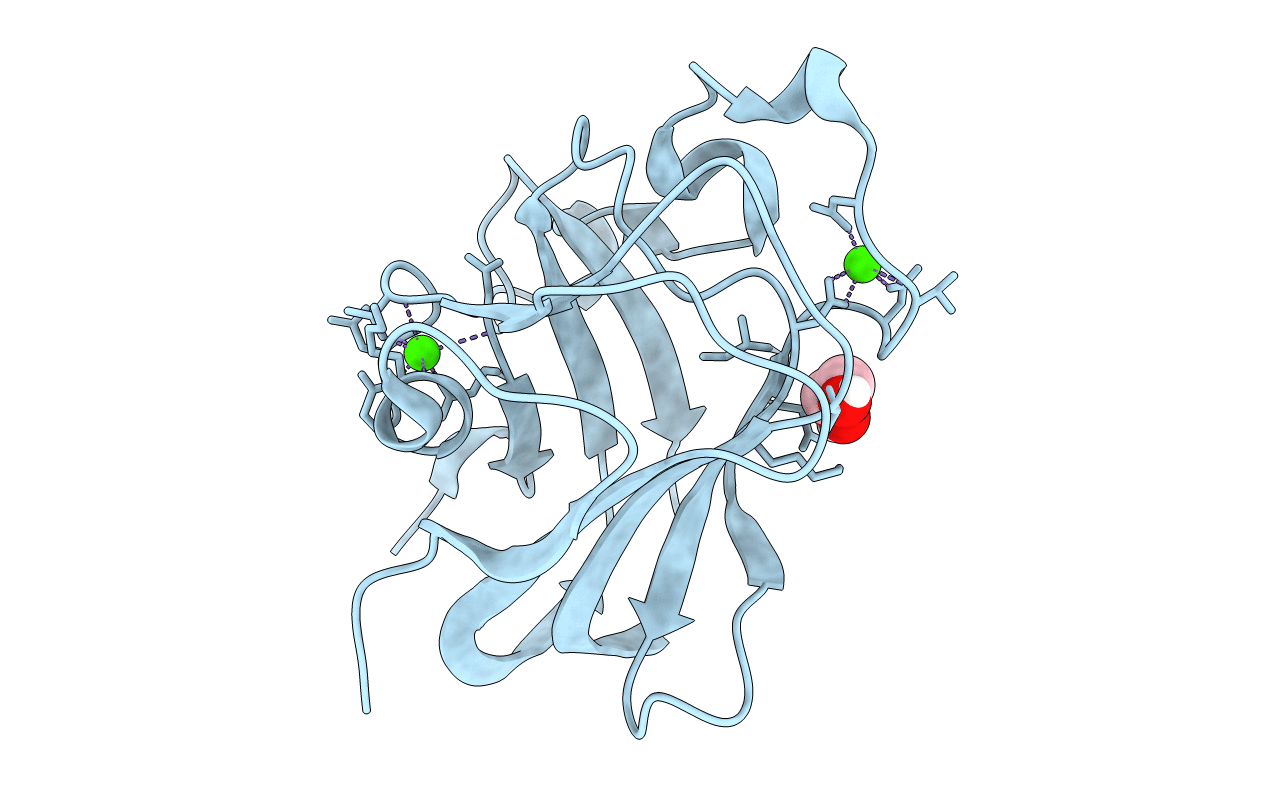 |
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2015-12-09 Classification: carbohydrate-binding module Ligands: CA, EDO |
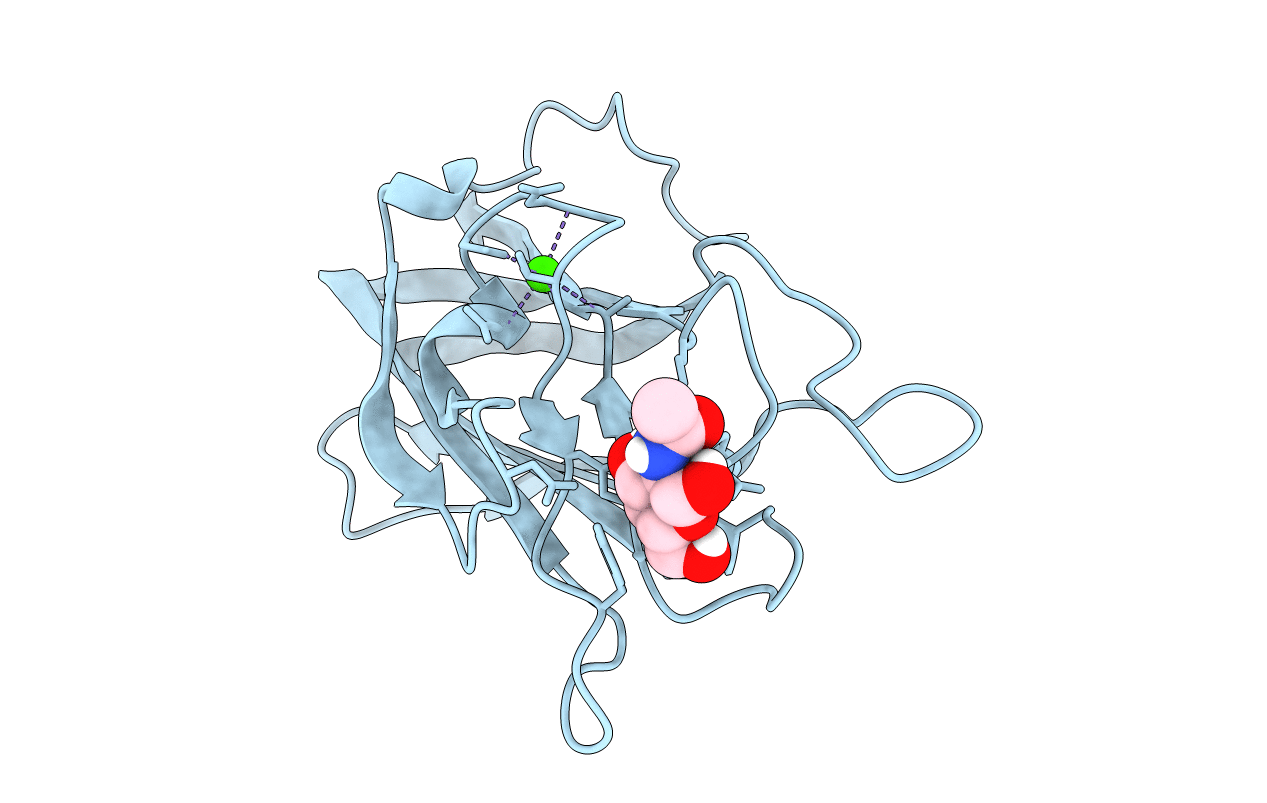 |
Structure Of Cbm32-3 From A Family 31 Glycoside Hydrolase From Clostridium Perfringens In Complex With N-Acetylgalactosamine
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-10-07 Classification: SUGAR BINDING PROTEIN Ligands: NGA, CA |
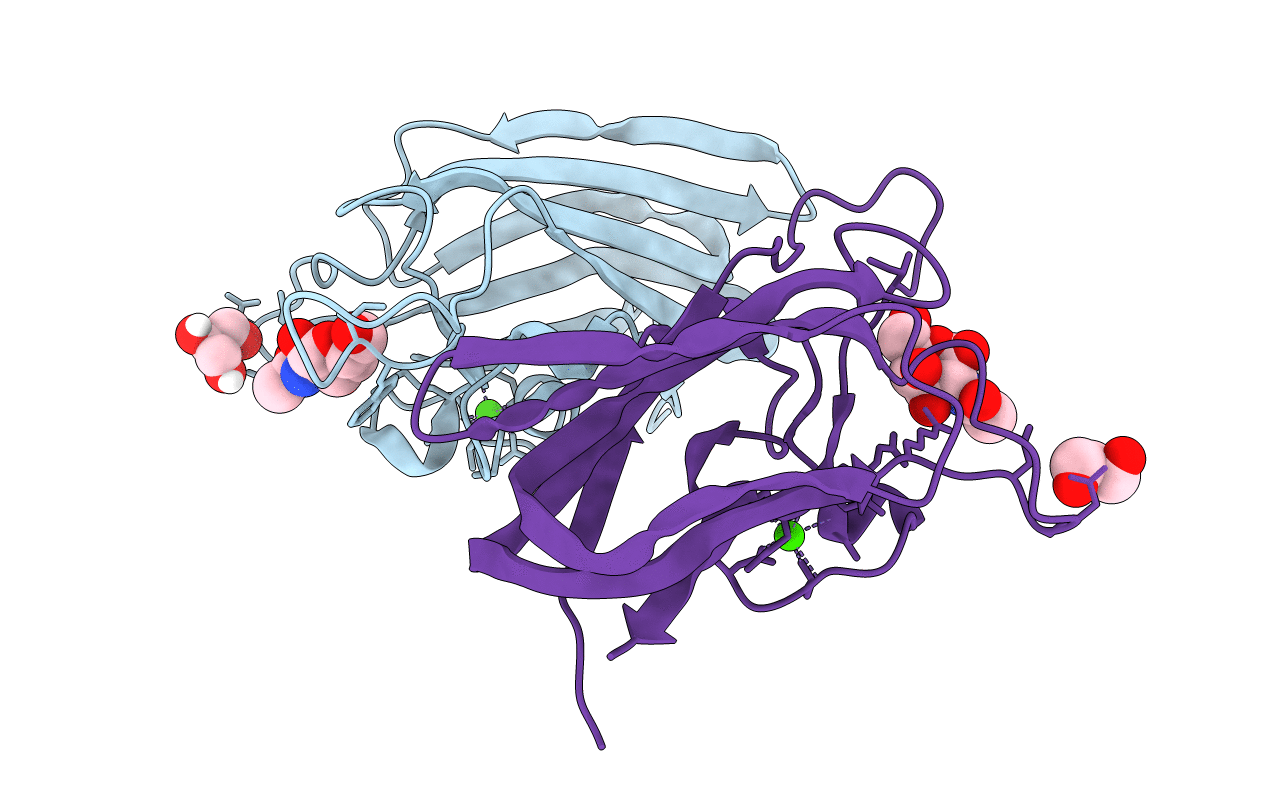 |
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-10-07 Classification: SUGAR BINDING PROTEIN Ligands: NGA, CA, GOL |
 |
Structure Of Cbm32-3 From A Family 31 Glycoside Hydrolase From Clostridium Perfringens In Complex With Galactose
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-12-24 Classification: SUGAR BINDING PROTEIN Ligands: CA, GAL |
 |
Structure Of Cbm32-3 From A Family 31 Glycoside Hydrolase From Clostridium Perfringens
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2014-07-23 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Structure Of Cbm32-1 From A Family 31 Glycoside Hydrolase From Clostridium Perfringens
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2014-07-16 Classification: SUGAR BINDING PROTEIN Ligands: CA, MG |
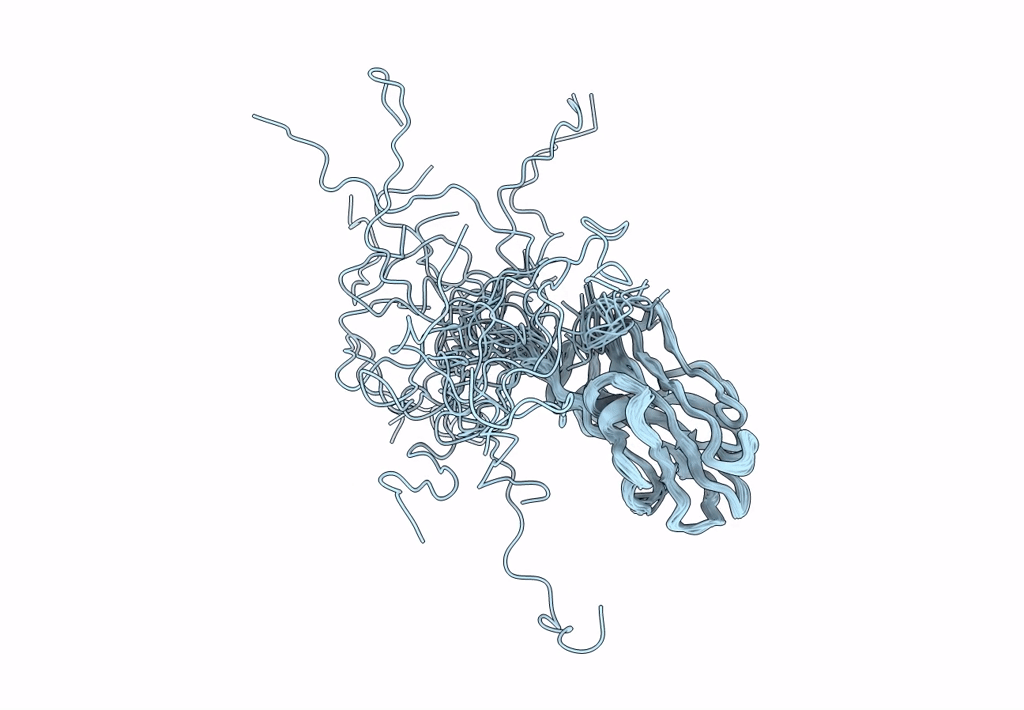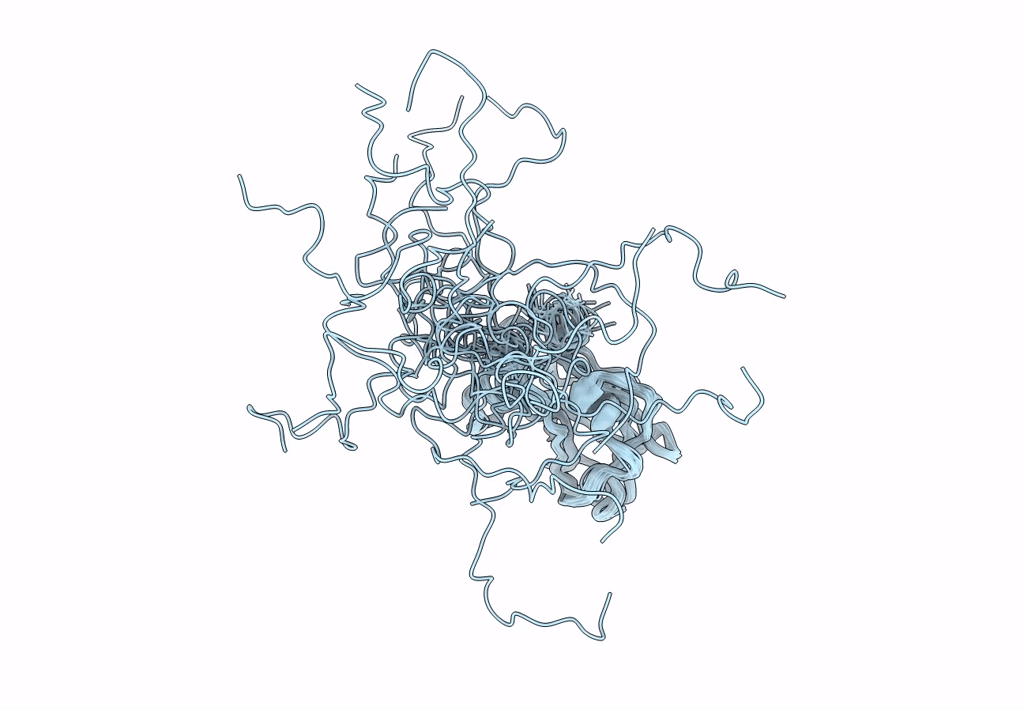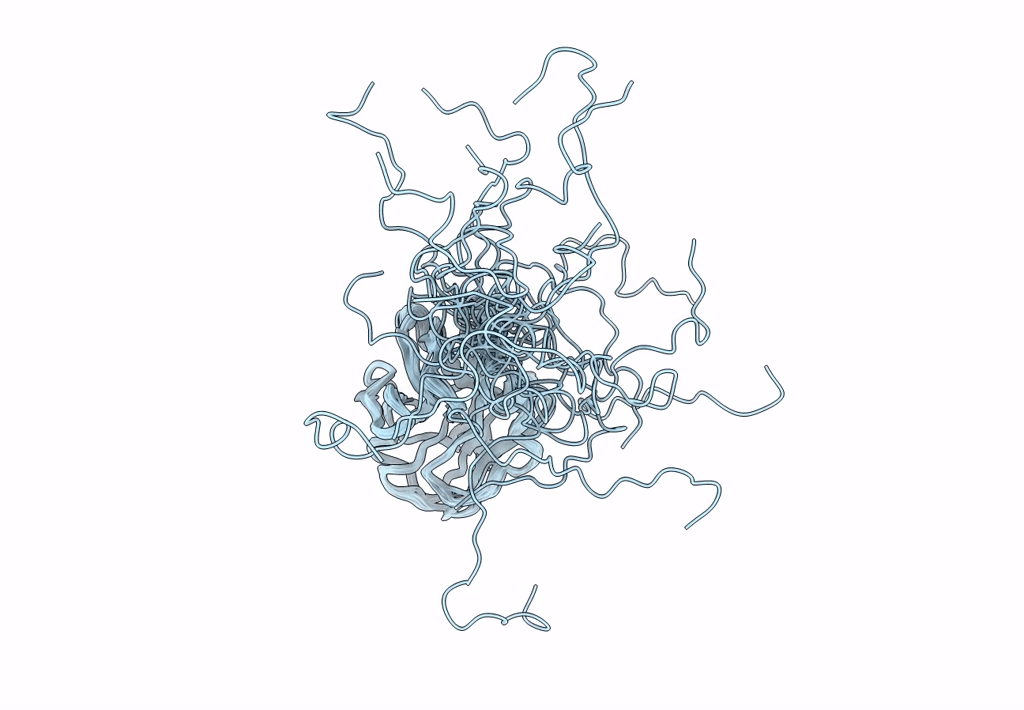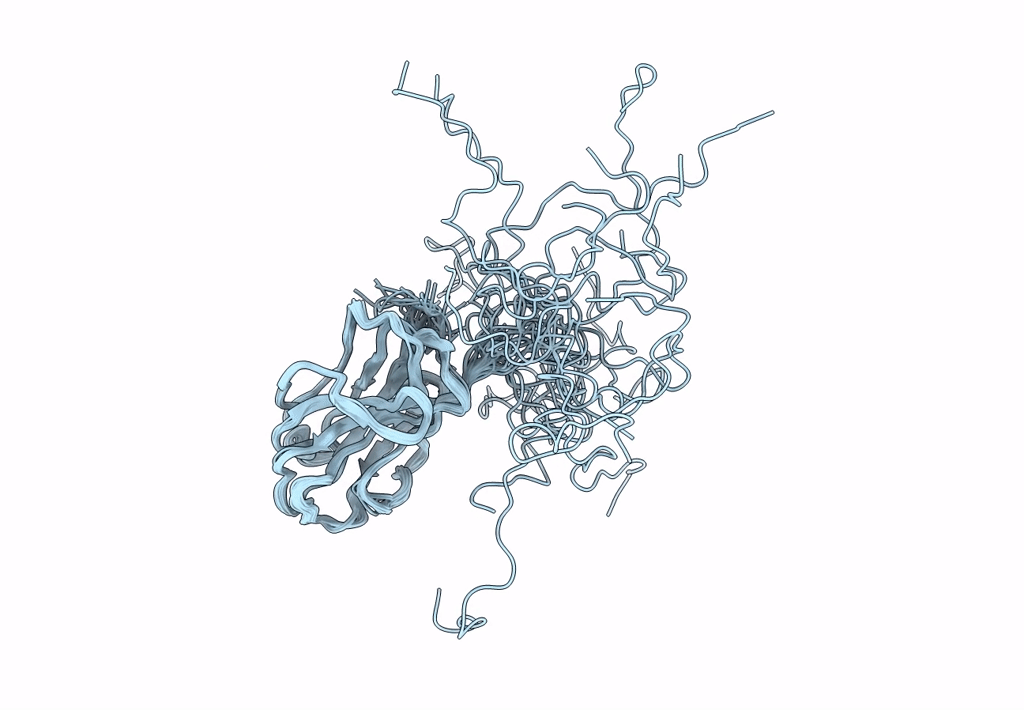 |
Solution Nmr Structure Of A Non-Canonical Galactose-Binding Cbm32 From Clostridium Perfringens
Organism: Clostridium perfringens
Method: SOLUTION NMR Release Date: 2013-05-01 Classification: HYDROLASE |

