Search Count: 142
 |
Organism: Clostridium cavendishii
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: LIGASE Ligands: ZN, MG |
 |
Organism: Yersinia ruckeri
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: METAL BINDING PROTEIN |
 |
Structure Of Yiua From Yersinia Ruckeri With Iron And Nitrilotriacetic Acid
Organism: Yersinia ruckeri
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: METAL BINDING PROTEIN Ligands: NTA, FE |
 |
Organism: Yersinia ruckeri
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
Organism: Yersinia ruckeri
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: METAL BINDING PROTEIN Ligands: FE, A1I15, SO4 |
 |
Organism: Kribbella flavida
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: NAP, SO4 |
 |
Imine Reductase Ir91 From Kribbella Flavida With Nadp+ And 5-Methoxy-2-Tetralone
Organism: Kribbella flavida
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: SO4, NAP, A1JDH, EDO |
 |
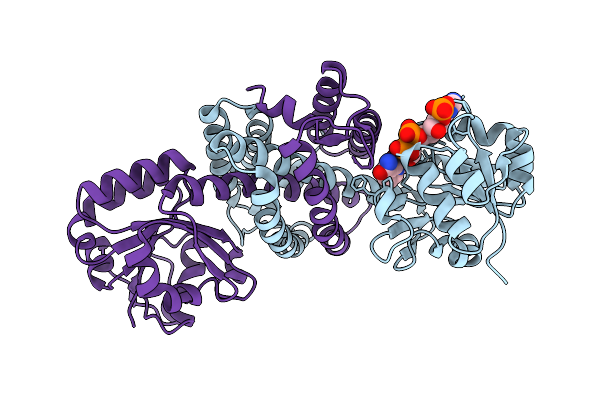
Imine Reductase Ir91 From Kribbella Flavida With Nadp+ And 5-Methoxy-(S)-2-(N-Methylamino)Tetralin
Organism: Kribbella flavida
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: SO4, NAP, A1JDG |
 |
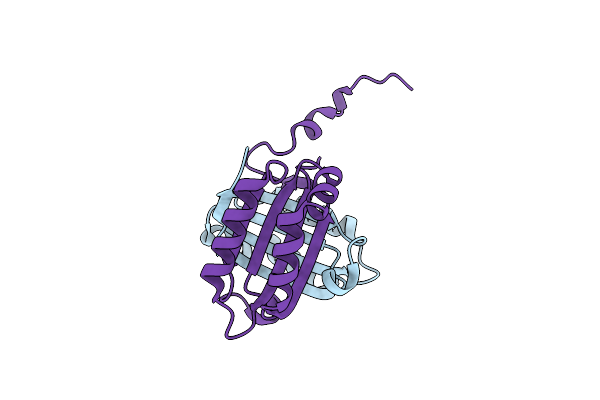
Structure Of Imine Reductase 361 From Micromonospora Sp. Mutant M125W/I127F/L179V/H250L
Organism: Micromonospora
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: OXIDOREDUCTASE Ligands: NAP |
 |
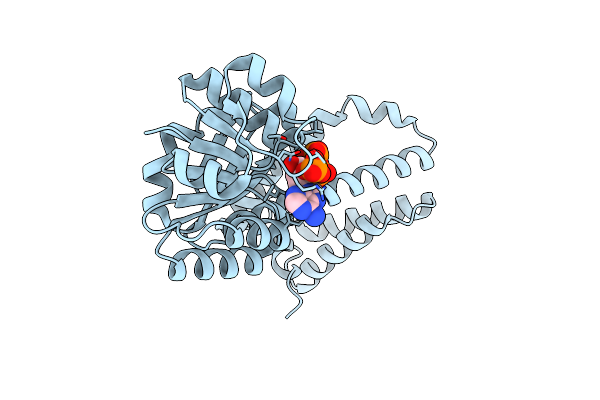
Organism: Brassica rapa
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2025-01-15 Classification: HYDROLASE |
 |
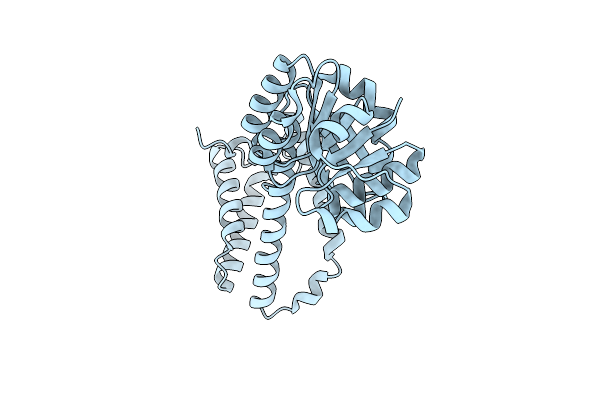
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-12-25 Classification: OXIDOREDUCTASE Ligands: ATR |
 |
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Release Date: 2024-12-25 Classification: OXIDOREDUCTASE |
 |
Nucleoside-2'-Deoxyribosyltransferase From Lactobacillus Leichmannii. Complex With Ribose And Cytosine
Organism: Lactobacillus leichmannii
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2024-12-25 Classification: TRANSFERASE Ligands: CYT, RIB |
 |
Nucleoside-2'-Deoxyribosyltransferase From Lactobacillus Leichmannii. Y7F/D72N Mutant With Cytidine
Organism: Lactobacillus leichmannii
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2024-12-25 Classification: TRANSFERASE Ligands: CTN |
 |
Nucleoside-2'-Deoxyribosyltransferase From Lactobacillus Leichmannii (Apo).
Organism: Lactobacillus leichmannii
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2024-12-04 Classification: TRANSFERASE |
 |
Nucleoside-2'-Deoxyribosyltransferase From Lactobacillus Leichmannii. Covalent Complex With 2-Deoxyribose.
Organism: Lactobacillus leichmannii
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2024-12-04 Classification: TRANSFERASE Ligands: 2DR |
 |
Nucleoside-2'-Deoxyribosyltransferase From Lactobacillus Leichmannii. Complex With 2-Deoxyribose, 7-Bromo-1H-Imidazo[4,5-B]Pyridine And 2'-Deoxycytidine
Organism: Lactobacillus leichmannii
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2024-12-04 Classification: TRANSFERASE Ligands: DCZ, A1H8P, 2DR |
 |
Amide Bond Synthetase From Streptomyces Hindustanus K492H Mutant In Complex With Amp-Cpp
Organism: Streptoalloteichus hindustanus
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2024-02-07 Classification: LIGASE Ligands: APC |
 |
Amide Bond Synthetase From Streptomyces Hindustanus K492H Mutant In Complex With Adenosine
Organism: Streptoalloteichus hindustanus
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2024-02-07 Classification: LIGASE Ligands: SO4, ADN |
 |
Organism: Streptoalloteichus hindustanus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-02-07 Classification: LIGASE Ligands: SO4 |

