Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
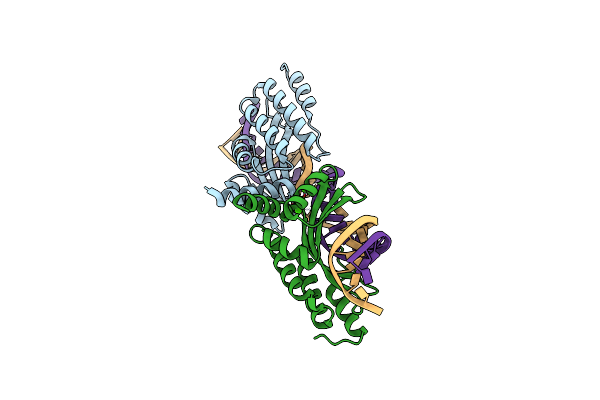 |
Crystal Structure Of The Homing Endonuclease I-Cvui In Complex With Its Target (Sro1.3) In The Presence Of 2 Mm Ca
Organism: Chlorella vulgaris, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-09-23 Classification: HYDROLASE/DNA Ligands: CA |
 |
Crystal Structure Of The Homing Endonuclease I-Cvui In Complex With Its Target (Sro1.3) In The Presence Of 2 Mm Mn
Organism: Chlorella vulgaris, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-09-23 Classification: HYDROLASE/DNA Ligands: MN |
 |
Crystal Structure Of The Homing Endonuclease I-Cvui In Complex With I- Crei Target (C1221) In The Presence Of 2 Mm Mg Revealing Dna Cleaved
Organism: Chlorella vulgaris, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-09-23 Classification: HYDROLASE/DNA Ligands: MG |
 |
Crystal Structure Of The Homing Endonuclease I-Cvui In Complex With I- Crei Target (C1221) In The Presence Of 2 Mm Mg Revealing Dna Not Cleaved
Organism: Chlorella vulgaris, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-09-23 Classification: HYDROLASE/DNA Ligands: MG |
 |
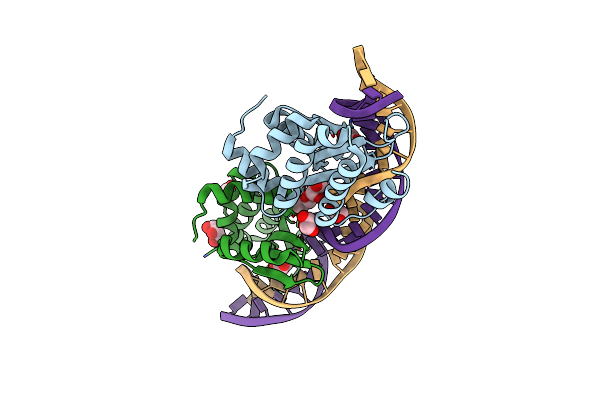
Crystal Structure Of The Mutant D75N I-Crei In Complex With Its Wild- Type Target (The Four Central Bases, 2Nn Region, Are Composed By Gtac From 5' To 3')
Organism: Chlamydomonas reinhardtii, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-05-02 Classification: HYDROLASE/DNA Ligands: PGO, MG, NA |
 |
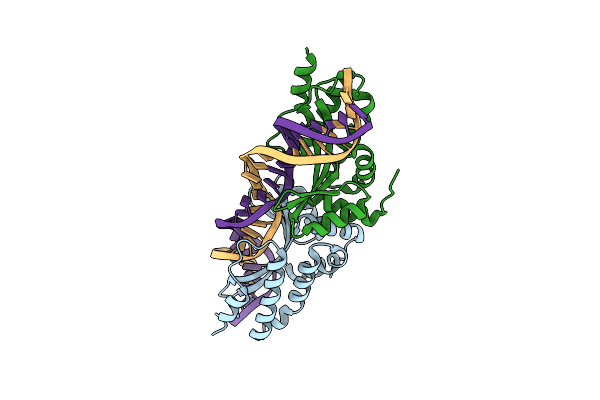
Crystal Structure Of The Mutant D75N I-Crei In Complex With Its Wild- Type Target In Absence Of Metal Ions At The Active Site (The Four Central Bases, 2Nn Region, Are Composed By Gtac From 5' To 3')
Organism: Chlamydomonas reinhardtii, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2012-05-02 Classification: HYDROLASE/DNA Ligands: GOL |
 |
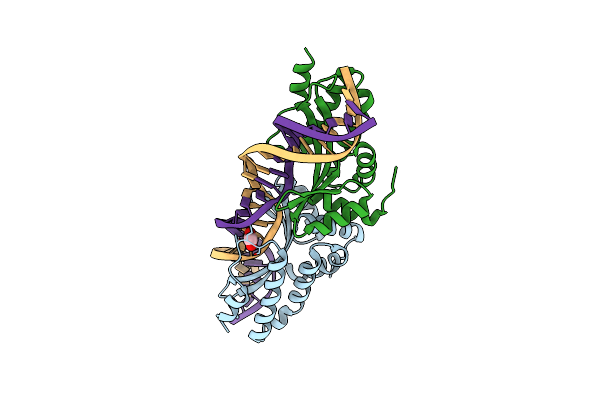
Crystal Structure Of The Mutant D75N I-Crei In Complex With An Altered Target (The Four Central Bases, 2Nn Region, Are Composed By Agcg From 5' To 3')
Organism: Chlamydomonas reinhardtii, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-05-02 Classification: HYDROLASE/DNA |
 |
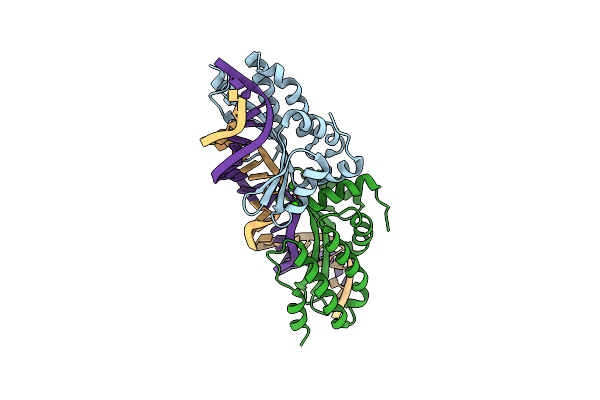
Crystal Structure Of The Mutant D75N I-Crei In Complex With An Altered Target (The Four Central Bases, 2Nn Region, Are Composed By Tgca From 5' To 3')
Organism: Chlamydomonas reinhardtii, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-05-02 Classification: HYDROLASE/DNA Ligands: EDO |
 |
Crystal Structure Of The Mutant D75N I-Crei In Complex With Its Wild- Type Target In Presence Of Ca At The Active Site (The Four Central Bases, 2Nn Region, Are Composed By Gtac From 5' To 3')
Organism: Chlamydomonas reinhardtii, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2012-05-02 Classification: HYDROLASE/DNA Ligands: CA |
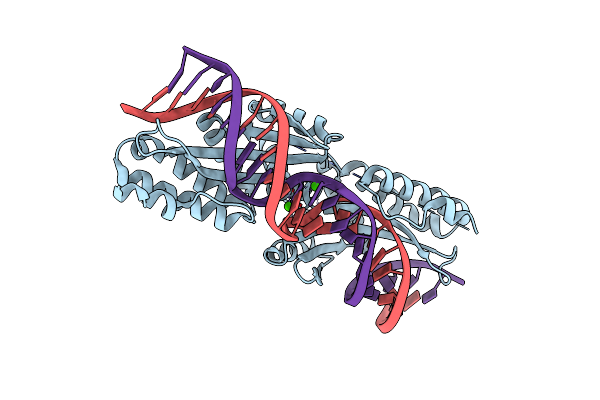 |
Molecular Basis Of Engineered Meganuclease Targeting Of The Endogenous Human Rag1 Locus
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2010-10-06 Classification: HYDROLASE/DNA Ligands: CA |
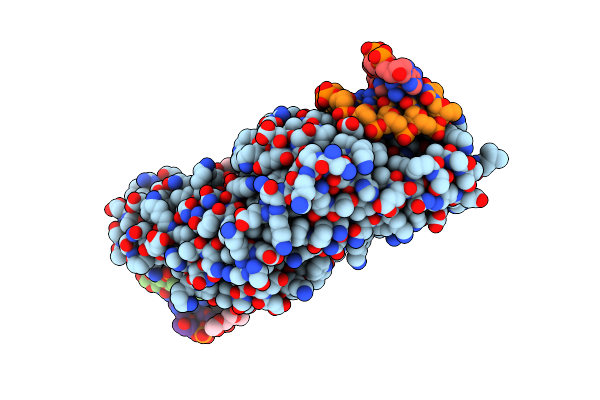 |
Molecular Basis Of Engineered Meganuclease Targeting Of The Endogenous Human Rag1 Locus
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-10-06 Classification: HYDROLASE/DNA Ligands: MN, EOH, 1PE |
 |
Molecular Basis Of Engineered Meganuclease Targeting Of The Endogenous Human Rag1 Locus
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-10-06 Classification: HYDROLASE/DNA Ligands: CA, PO4 |
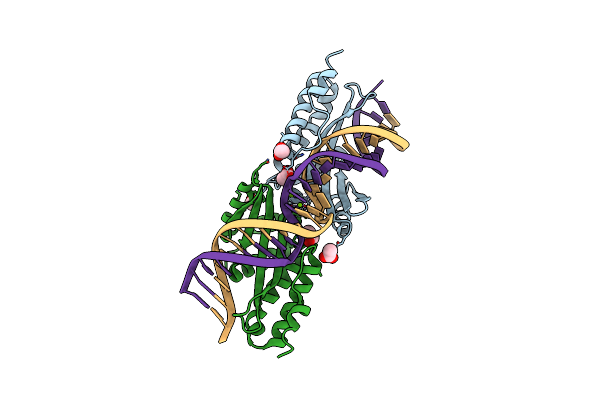 |
Molecular Basis Of Engineered Meganuclease Targeting Of The Endogenous Human Rag1 Locus
Organism: Chlamydomonas reinhardtii, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2010-09-29 Classification: DNA BINDING PROTEIN/DNA Ligands: ACT, PGO, MG |
 |
Organism: Desulfurococcus mobilis
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2008-11-11 Classification: DNA BINDING PROTEIN Ligands: CA, ACT |
 |
Organism: Desulfurococcus mobilis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-11-11 Classification: DNA BINDING PROTEIN Ligands: MN, ACT |
 |
Organism: Escherichia coli o6
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2007-05-22 Classification: TRANSFERASE Ligands: ADP |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2007-05-22 Classification: TRANSFERASE Ligands: AFH |
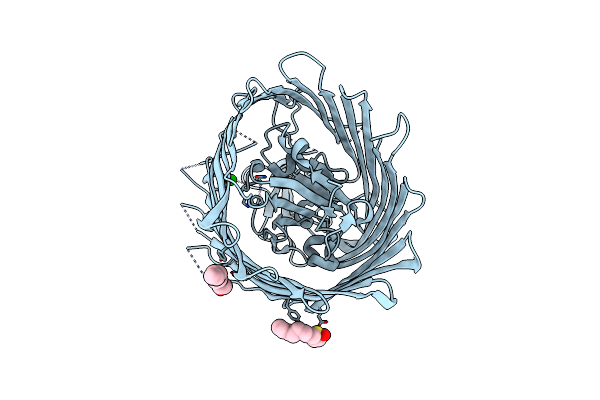 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2007-05-08 Classification: PROTEIN TRANSPORT Ligands: SR, OES |
 |
Crystal Structure Of The Colicin I Receptor Cir From E.Coli In Complex With Receptor Binding Domain Of Colicin Ia.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-05-08 Classification: Protein Transport,Antimicrobial Protein Ligands: LDA |
 |
Organism: Escherichia coli uti89
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-05-01 Classification: TRANSFERASE |