Search Count: 35
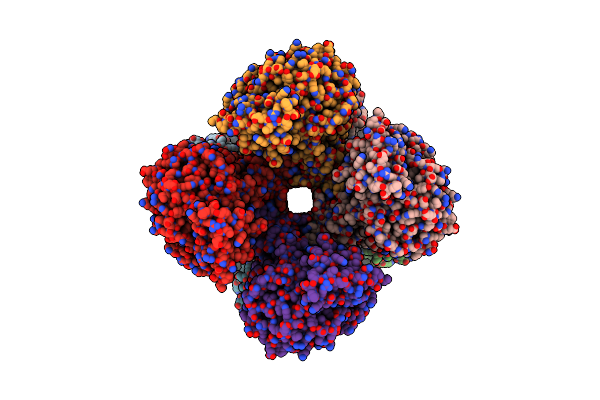 |
Cryoem Structure Of Aspergillus Nidulans Utp-Glucose-1-Phosphate Uridylyltransferase
Organism: Aspergillus nidulans fgsc a4
Method: ELECTRON MICROSCOPY Release Date: 2023-06-28 Classification: SUGAR BINDING PROTEIN |
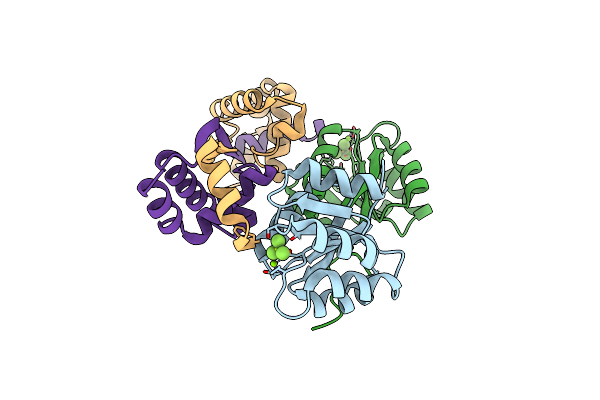 |
Organism: Methanococcus maripaludis x1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-04-27 Classification: METAL BINDING PROTEIN Ligands: MG, BEF |
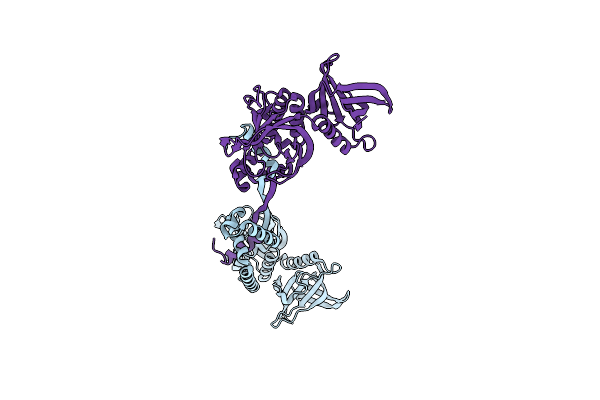 |
Organism: Pyrococcus horikoshii (strain atcc 700860 / dsm 12428 / jcm 9974 / nbrc 100139 / ot-3)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2022-04-27 Classification: METAL BINDING PROTEIN |
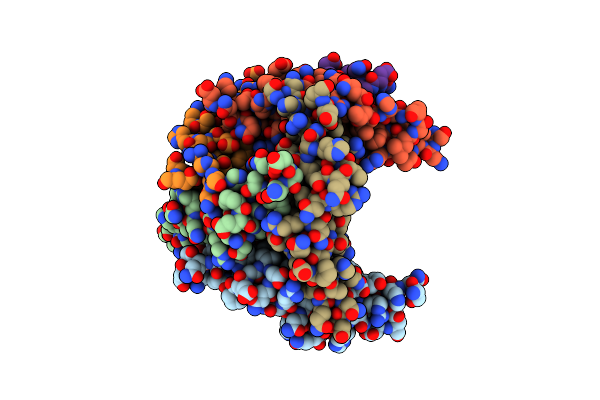 |
Organism: Streptomyces davaonensis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-05-13 Classification: FLAVOPROTEIN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-12-04 Classification: TRANSFERASE Ligands: OCA, CO8 |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2019-11-06 Classification: BIOSYNTHETIC PROTEIN Ligands: NDP, FMN |
 |
Organism: Streptomyces coelicolor (strain atcc baa-471 / a3(2) / m145)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-03-27 Classification: FLAVOPROTEIN Ligands: FMN, COA, CL, NA, SO4 |
 |
Organism: Pyrococcus horikoshii (strain atcc 700860 / dsm 12428 / jcm 9974 / nbrc 100139 / ot-3)
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2018-12-12 Classification: SIGNALING PROTEIN |
 |
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2018-10-31 Classification: SIGNALING PROTEIN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-02-21 Classification: TRANSFERASE Ligands: COA |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2018-01-24 Classification: TRANSFERASE Ligands: COA, MLC |
 |
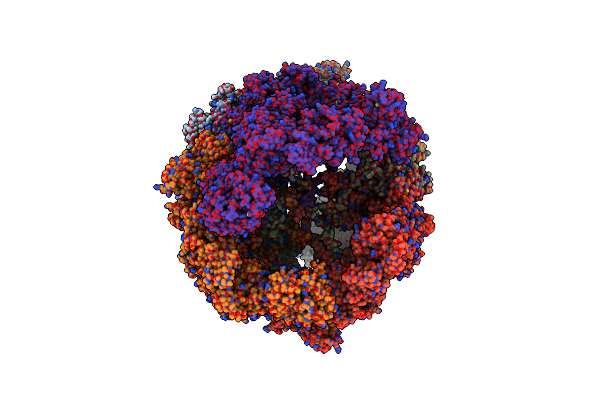
Structure And Conformational Variability Of The Mycobacterium Tuberculosis Fatty Acid Synthase Multienzyme Complex
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Resolution:20.00 Å Release Date: 2014-07-09 Classification: HYDROLASE Ligands: FMN |
 |
Structure And Conformational Variability Of The Mycobacterium Tuberculosis Fatty Acid Synthase Multienzyme Complex
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Resolution:17.50 Å Release Date: 2014-07-09 Classification: HYDROLASE Ligands: FMN |
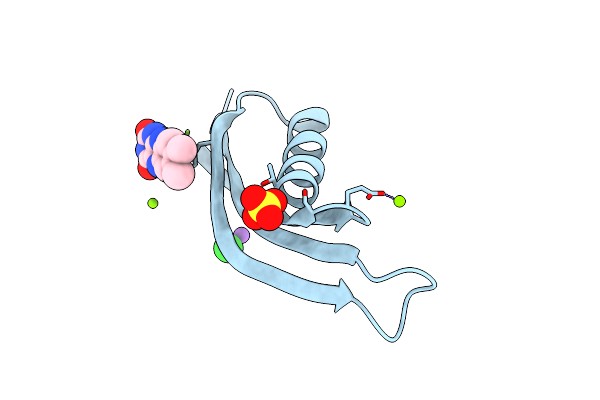 |
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-07-17 Classification: FLAVOPROTEIN Ligands: MG, NA, CL, SO4, RBF |
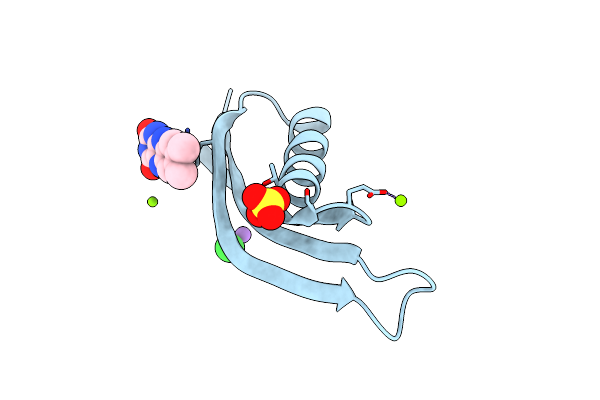 |
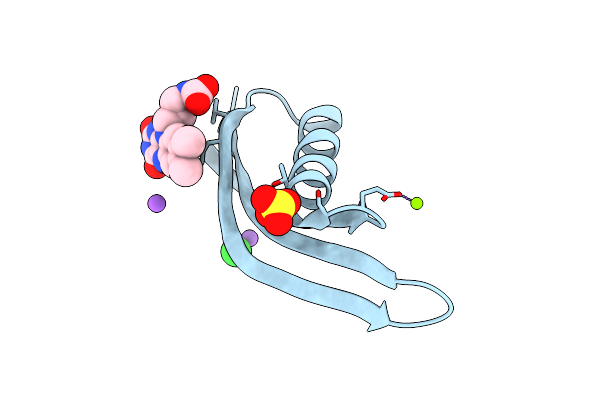
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-07-17 Classification: FLAVOPROTEIN Ligands: MG, NA, CL, SO4, RBF |
 |
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-05-29 Classification: FLAVOPROTEIN |
 |
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-05-29 Classification: FLAVOPROTEIN Ligands: MG, NA, CL, SO4, RBF |
 |
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2010-05-19 Classification: HYDROLASE Ligands: PO4, ZN, MG, NA, CL |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2009-09-01 Classification: TRANSFERASE Ligands: CER, FMN |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2009-08-25 Classification: TRANSFERASE |

