Search Count: 28
 |
 |
M. Tuberculosis Meets European Lead Factory: Identification And Structural Characterization Of Novel Rv0183 Inhibitors Using X-Ray Crystallography: Elf5
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: HYDROLASE Ligands: 8KE, I3F, CL |
 |
M. Tuberculosis Meets European Lead Factory: Identification And Structural Characterization Of Novel Rv0183 Inhibitors Using X-Ray Crystallography: Elf8
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: HYDROLASE Ligands: 8D5, MPD, EDO, PGE, PEG |
 |
M. Tuberculosis Meets European Lead Factory: Identification And Structural Characterization Of Novel Rv0183 Inhibitors Using X-Ray Crystallography: Elf1
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: HYDROLASE Ligands: 7WW, PO4, MPD, DMS, EOH, EDO |
 |
Self Assembly Domain Of The Surface Layer Protein Of Viridibacillus Arvi (Aa 765-844)
Organism: Viridibacillus arvi
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-08-28 Classification: STRUCTURAL PROTEIN Ligands: BR |
 |
Cell Wall Anchoring Domain Of The Surface Layer Protein Of Methanococcus Voltae (Aa 24-75; 484-576)
Organism: Methanococcus voltae
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-08-28 Classification: STRUCTURAL PROTEIN |
 |
The Crystal Structure Of 2-Hydroxy-3-Keto-Glucal Hydratase Athyd From A. Tumefaciens
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2024-08-07 Classification: LYASE Ligands: MN |
 |
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2024-08-07 Classification: LYASE Ligands: GOL, EDO, CA, A1H2X |
 |
Crystal Structure Of Slpa Domain Ii (Aa 201-310), Domain That Is Involved In The Self-Assembly And Dimerization Of The S-Layer From Lactobacillus Acidophilus
Organism: Lactobacillus acidophilus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-09-06 Classification: STRUCTURAL PROTEIN Ligands: NCA |
 |
S-Layer Protein Slpa From Lactobacillus Amylovorus, Domain I (Aa 32-209), Important For Self-Assembly
Organism: Lactobacillus amylovorus
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2023-09-06 Classification: STRUCTURAL PROTEIN Ligands: PO4 |
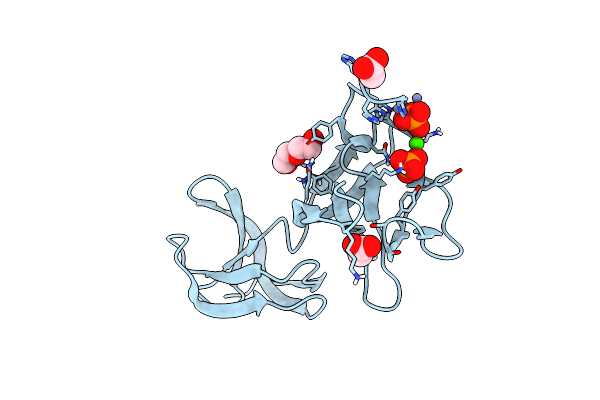 |
Crystal Structure Of S-Layer Protein Slpx From Lactobacillus Acidophilus, Domain Iii (Aa 363-499)
Organism: Lactobacillus acidophilus atcc 4796
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-08-23 Classification: CELL ADHESION Ligands: ZN, PO4, CA, ACT, PEG |
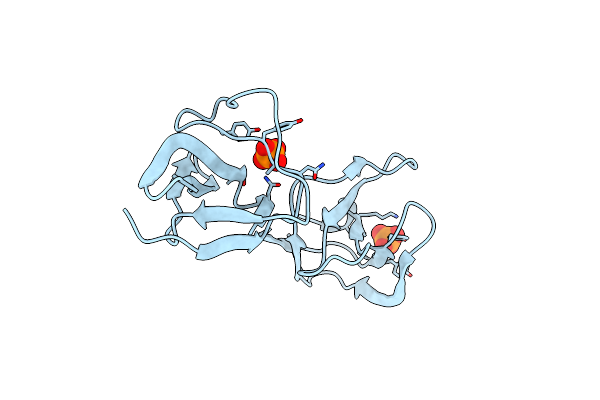 |
Crystal Structure Of The Teichoic Acid Binding Domain Of Slpa, S-Layer Protein From Lactobacillus Acidophilus (Aa. 314-444)
Organism: Lactobacillus acidophilus
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2023-08-16 Classification: CELL ADHESION Ligands: PO4 |
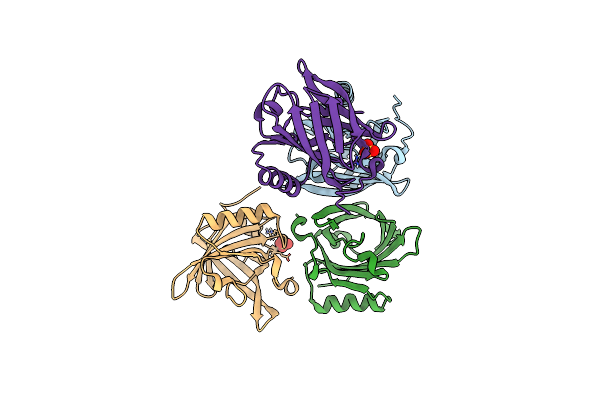 |
Organism: Felis catus
Method: X-RAY DIFFRACTION Release Date: 2023-08-16 Classification: ALLERGEN Ligands: SO4 |
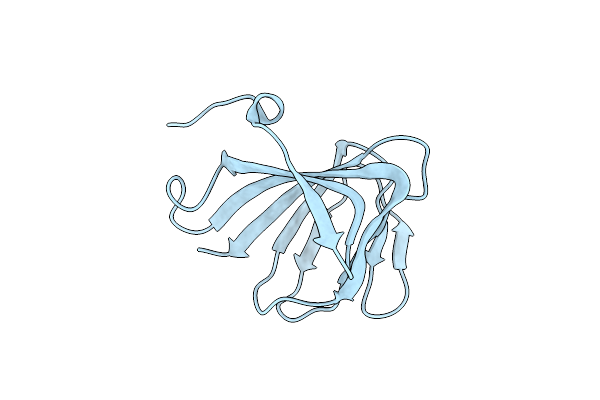 |
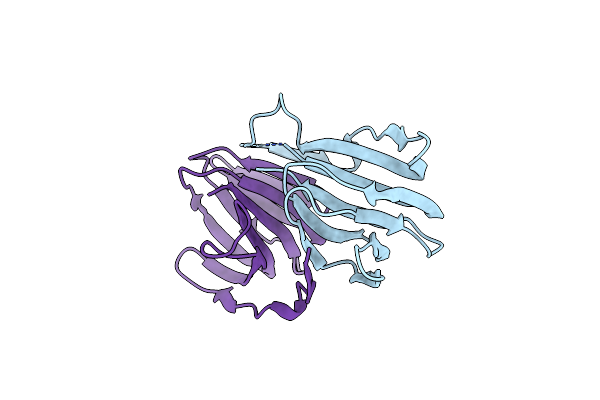
Crystal Structure Of A Manganese-Containing Cupin (Tm1459) From Thermotoga Maritima, Variant C106Q
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2023-03-08 Classification: METAL BINDING PROTEIN Ligands: CL |
 |
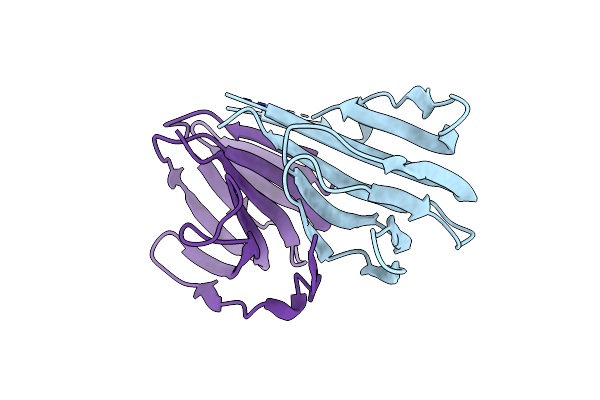
Crystal Structure Of A Manganese-Containing Cupin (Tm1459) From Thermotoga Maritima, Variant Aifq (Y7A/M38I/Y83F/C106Q)
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-03-08 Classification: METAL BINDING PROTEIN |
 |
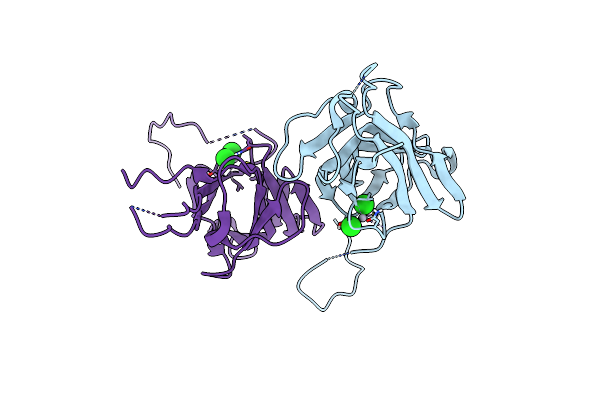
Crystal Structure Of A Manganese-Containing Cupin (Tm1459) From Thermotoga Maritima, Variant 208 (V19I/R23H/M38I/I60F/C106Q)
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-03-08 Classification: METAL BINDING PROTEIN |
 |
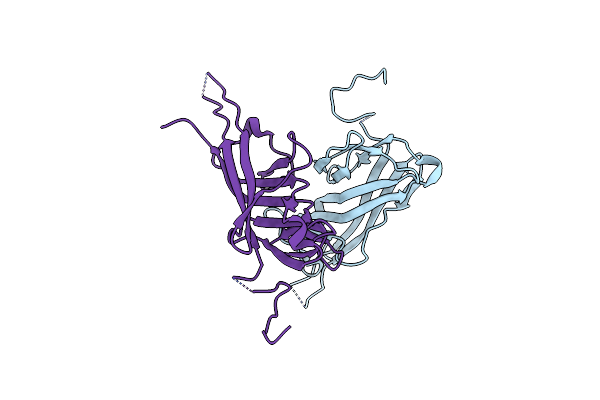
Crystal Structure Of S-Layer Protein Slpa From Lactobacillus Acidophilus, Domain I, Co-Crystallization With Hgcl2, Mutation Ser146Cys, (Aa 32-198)
Organism: Lactobacillus acidophilus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-12-28 Classification: STRUCTURAL PROTEIN Ligands: HG, CL |
 |
Crystal Structure Of S-Layer Protein Slpa From Lactobacillus Acidophilus, Domain I (Aa 32-198)
Organism: Lactobacillus acidophilus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-12-28 Classification: STRUCTURAL PROTEIN |
 |
Crystal Structure Of S-Layer Protein Slpa From Lactobacillus Amylovorus, Domain I (Aa 48-213)
Organism: Lactobacillus amylovorus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-12-28 Classification: STRUCTURAL PROTEIN Ligands: PO4, NA |
 |
Crystal Structure Of S-Layer Protein Slpa From Lactobacillus Acidophilus, Domain Iii (Aa 309-444)
Organism: Lactobacillus acidophilus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-12-21 Classification: CELL ADHESION Ligands: CL, BTB |

