Search Count: 142
 |
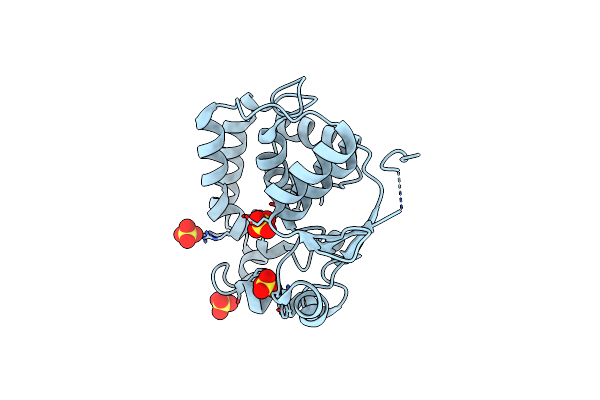
Crystal Structure Of The Substrate Binding Domain Protein Of The Abc Transporter Pbp2_Yxem From Vibrio Cholerae
Organism: Vibrio cholerae o1 biovar el tor str. n16961
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-08-28 Classification: TRANSPORT PROTEIN Ligands: PGO, CL, K, PGR, SO4 |
 |
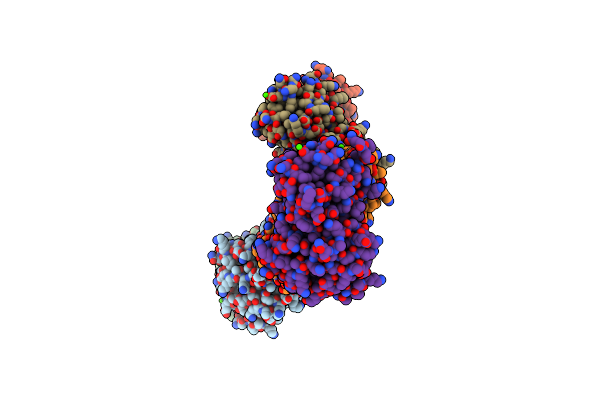
Organism: Francisella tularensis subsp. tularensis schu s4
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2022-01-05 Classification: TRANSPORT PROTEIN Ligands: SO4, CL |
 |
Crystal Structure Of The Pbp2_Yvgl_Like Protein Lmo1041 From Listeria Monocytogene
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2021-12-29 Classification: TRANSPORT PROTEIN Ligands: GOL, CA, SO4, CL |
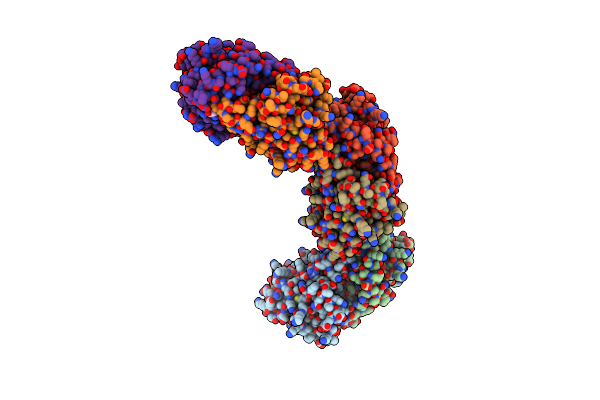 |
2.1 Angstrom Resolution Crystal Structure Of 5'-Methylthioadenosine/S-Adenosylhomocysteine Nucleosidase (Mtnn) From Haemophilus Influenzae Pittii.
Organism: Haemophilus influenzae pittii
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-07-17 Classification: HYDROLASE Ligands: HCS, ADE, BO3 |
 |
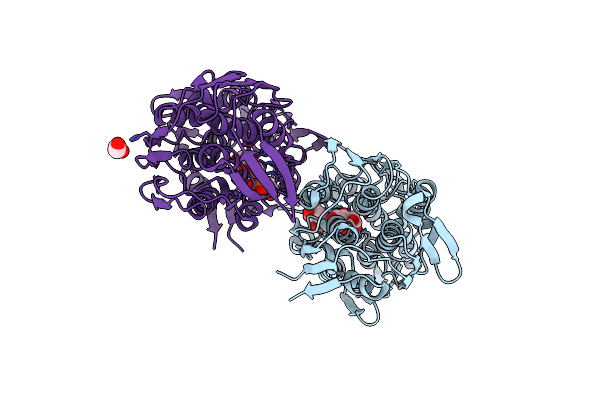
Crystal Structure Of Adenosine Deaminase From Salmonella Typhimurium With Pentostatin (Deoxycoformycin)
Organism: Salmonella typhimurium (strain lt2 / sgsc1412 / atcc 700720)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2019-02-06 Classification: HYDROLASE Ligands: DCF, ZN, CA, FMT |
 |
1.3 Angstrom Resolution Crystal Structure Of Udp-N-Acetylglucosamine 1-Carboxyvinyltransferase From Streptococcus Pneumoniae In Complex With (2R)-2-(Phosphonooxy)Propanoic Acid.
Organism: Streptococcus pneumoniae serotype 4 (strain atcc baa-334 / tigr4)
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: 0V5, CIT, CL, EDO |
 |
1.7 Angstrom Resolution Crystal Structure Of Arginase From Bacillus Subtilis Subsp. Subtilis Str. 168
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-01-02 Classification: HYDROLASE Ligands: CL, NA, MG, FMT, URE, EDO, SO4, PEG, GOA |
 |
1.50 Angstrom Resolution Crystal Structure Of Argininosuccinate Synthase From Bordetella Pertussis In Complex With Amp.
Organism: Bordetella pertussis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-08-01 Classification: HYDROLASE Ligands: AMP, CL, SO4, EDO, MLA |
 |
Crystal Structure Of Bifunctional Enzyme Fold-Methylenetetrahydrofolate Dehydrogenase/Cyclohydrolase In The Complex With Methotrexate From Campylobacter Jejuni
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-05-23 Classification: OXIDOREDUCTASE Ligands: GUN, K, GOL, CL, TRS, MTX, DTT |
 |
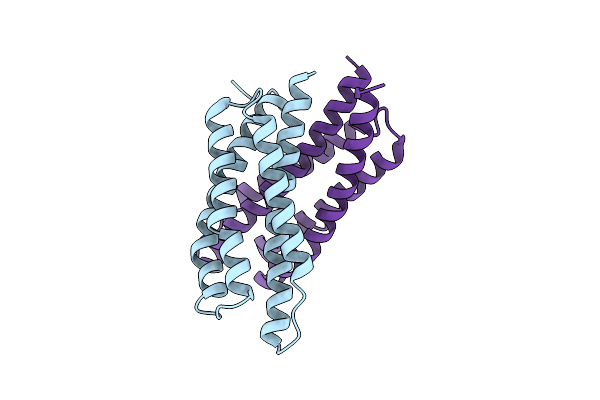
Organism: Haemophilus influenzae (strain atcc 51907 / dsm 11121 / kw20 / rd)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2018-05-23 Classification: LIGASE Ligands: TRP |
 |
2.0 Angstrom Resolution Crystal Structure Of N-Terminal Ligand-Binding Domain Of Putative Methyl-Accepting Chemotaxis Protein From Salmonella Enterica
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. lt2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-05-16 Classification: TRANSFERASE Ligands: CL |
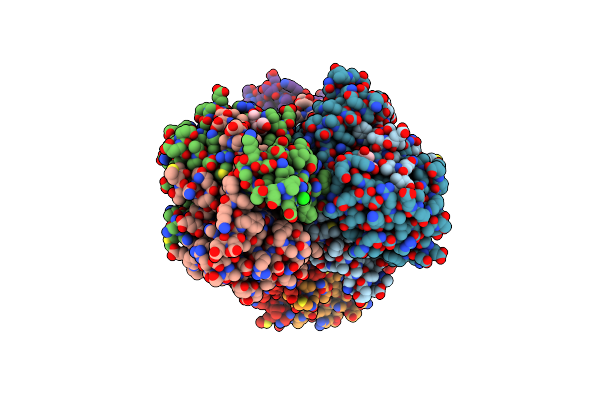 |
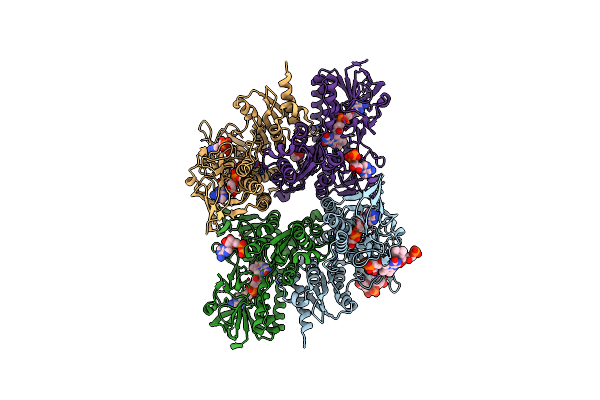
2.2 Angstrom Resolution Crystal Structure Oxygen-Insensitive Nad(P)H-Dependent Nitroreductase Nfsb From Vibrio Vulnificus In Complex With Fmn
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2018-04-25 Classification: OXIDOREDUCTASE Ligands: FMN, GOL, PEG, CL, PGE |
 |
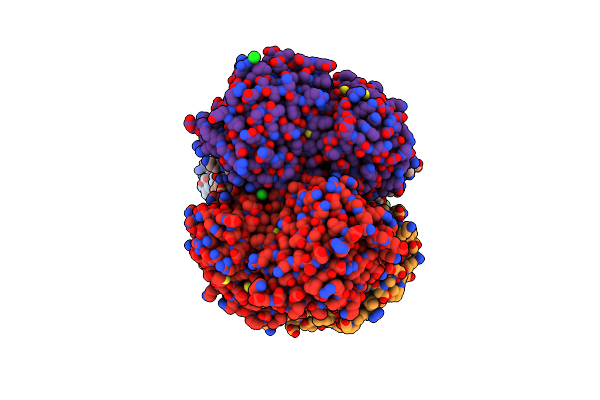
2.3 Angstrom Resolution Crystal Structure Of Dihydrolipoamide Dehydrogenase From Burkholderia Cenocepacia In Complex With Fad And Nad
Organism: Burkholderia cenocepacia (strain atcc baa-245 / dsm 16553 / lmg 16656 / nctc 13227 / j2315 / cf5610)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-03-21 Classification: OXIDOREDUCTASE Ligands: CL, FAD, NAD, FMN, ADP, MLT |
 |
2.75 Angstrom Resolution Crystal Structure Of Udp-N-Acetylglucosamine 1-Carboxyvinyltransferase From Pseudomonas Putida In Complex With Uridine-Diphosphate-2(N-Acetylglucosaminyl) Butyric Acid, (2R)-2-(Phosphonooxy)Propanoic Acid And Magnesium
Organism: Pseudomonas putida (strain atcc 47054 / dsm 6125 / ncimb 11950 / kt2440)
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2018-03-21 Classification: TRANSFERASE Ligands: EPU, 0V5, MG, CL |
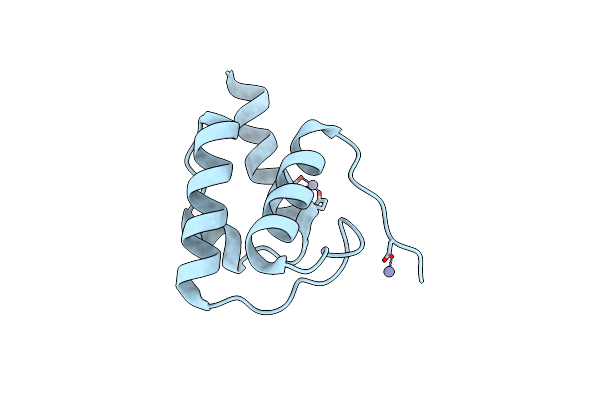 |
1.9 Angstrom Resolution Crystal Structure Of Acyl Carrier Protein Domain (Residues 1350-1461) Of Polyketide Synthase Pks13 From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-01-31 Classification: TRANSPORT PROTEIN Ligands: ZN |
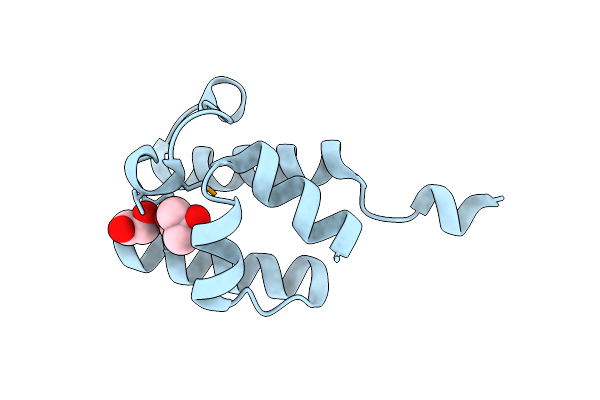 |
1.16 Angstrom Resolution Crystal Structure Of Acyl Carrier Protein Domain (Residues 1-100) Of Polyketide Synthase Pks13 From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2018-01-24 Classification: TRANSPORT PROTEIN Ligands: MPD, EDO |
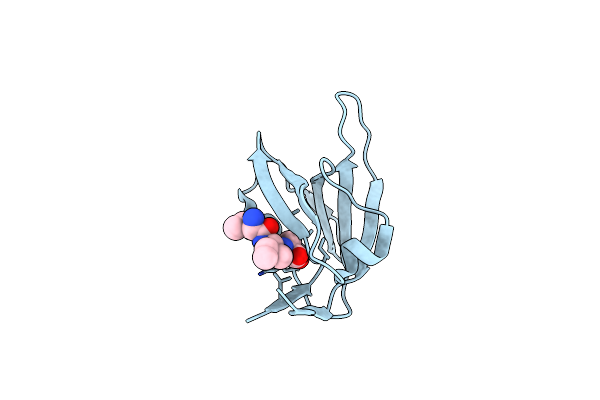 |
1.45 Angstrom Resolution Crystal Structure Of Pdz Domain Of Carboxy-Terminal Protease From Vibrio Cholerae In Complex With Peptide.
Organism: Vibrio cholerae serotype o1, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2018-01-03 Classification: HYDROLASE/PEPTIDE Ligands: CL, IOD |
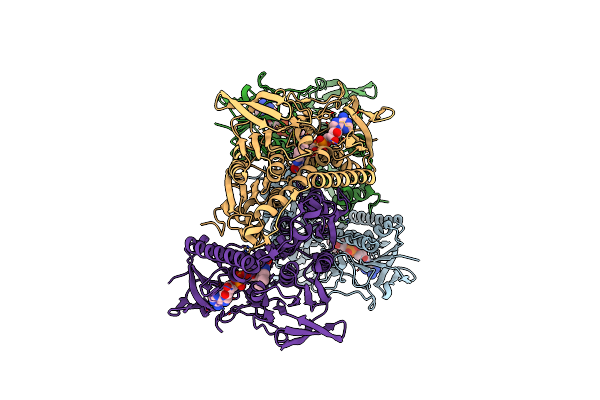 |
1.83 Angstrom Resolution Crystal Structure Of Dihydrolipoyl Dehydrogenase From Acinetobacter Baumannii In Complex With Fad.
Organism: Acinetobacter baumannii (strain atcc 19606 / dsm 30007 / cip 70.34 / jcm 6841 / nbrc 109757 / ncimb 12457 / nctc 12156 / 81)
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2018-01-03 Classification: OXIDOREDUCTASE Ligands: FAD, CL, MG |
 |
2.45 Angstrom Resolution Crystal Structure Thioredoxin Reductase From Francisella Tularensis.
Organism: Francisella tularensis subsp. tularensis (strain schu s4 / schu 4)
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2017-12-27 Classification: OXIDOREDUCTASE Ligands: CL, SO4 |
 |
2.55 Angstrom Resolution Crystal Structure Of N-Terminal Fragment (Residues 1-493) Of Dna Topoisomerase Iv Subunit A From Pseudomonas Putida
Organism: Pseudomonas putida (strain atcc 47054 / dsm 6125 / ncimb 11950 / kt2440)
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2017-12-06 Classification: ISOMERASE |

