Search Count: 12
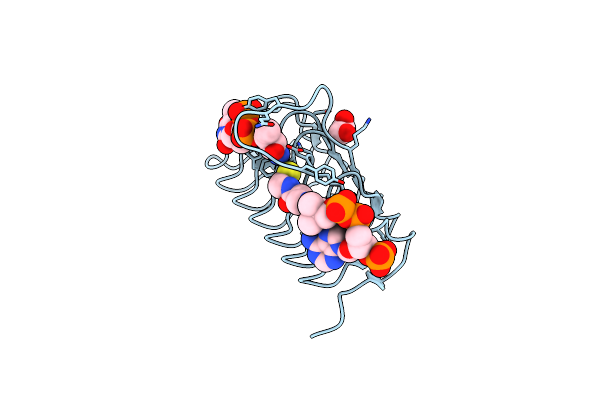 |
Crystal Structure Of An N-Acetyltransferase From Helicobacter Pullorum In The Presence Of Coenzyme A And Dtdp-3-Amino-3,6-Dideoxy-D-Glucose
Organism: Helicobacter pullorum
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-09-22 Classification: TRANSFERASE Ligands: COA, T3Q, EDO |
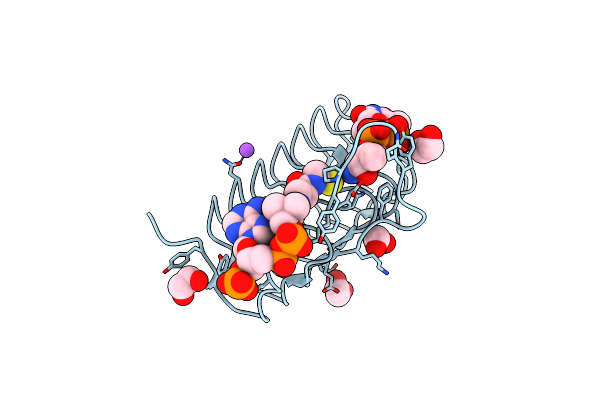 |
Crystal Structure Of An N-Acetyltransferase From Helicobacter Pullorum In The Presence Of Coenzyme A And Dtdp-3-Amino-3,6-Dideoxy-D-Galactose
Organism: Helicobacter pullorum
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2021-09-22 Classification: TRANSFERASE Ligands: T3F, CAO, EDO, NA |
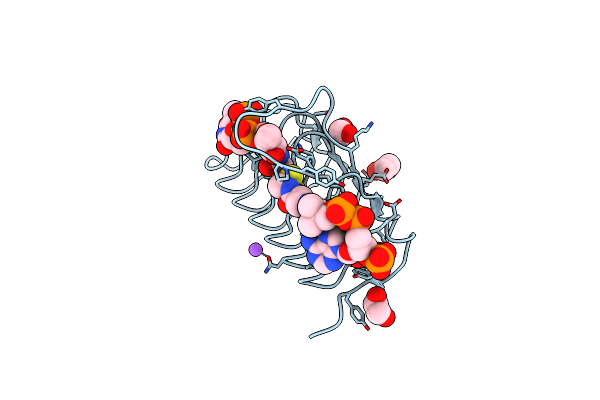 |
Crystal Structure Of An N-Acetyltransferase From Helicobacter Pullorum In The Presence Of Coenzyme A And Dtdp-3-Acetamido-3,6-Dideoxy-D-Glucose
Organism: Helicobacter pullorum
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-09-22 Classification: TRANSFERASE Ligands: 87N, COA, EDO, NA |
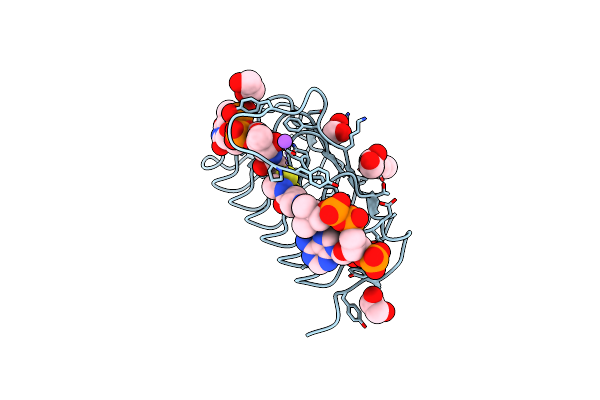 |
Crystal Structure Of An N-Acetyltransferase From Helicobacter Pullorum In The Presence Of Coenzyme A And Dtdp-3-Acetamido-3,6-Dideoxy-D-Galactose
Organism: Helicobacter pullorum
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2021-09-22 Classification: TRANSFERASE Ligands: 87Z, COA, EDO, NA |
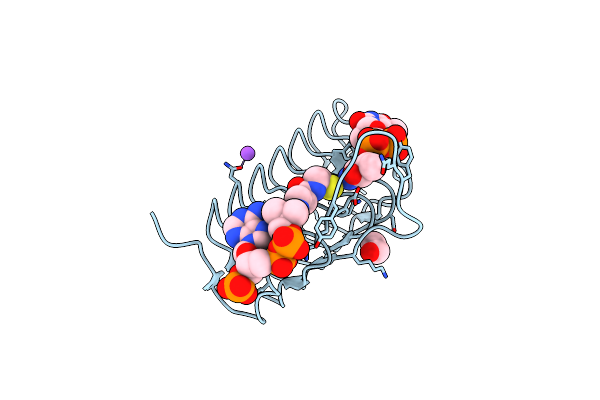 |
Crystal Structure Of An N-Acetyltransferase, C80T Mutant, From Helicobacter Pullorum In The Presence Of Coenzyme A And Dtdp-3-Amino-3,6-Dideoxy-D-Glucose
Organism: Helicobacter pullorum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-09-22 Classification: TRANSFERASE Ligands: T3Q, COA, EDO, NA |
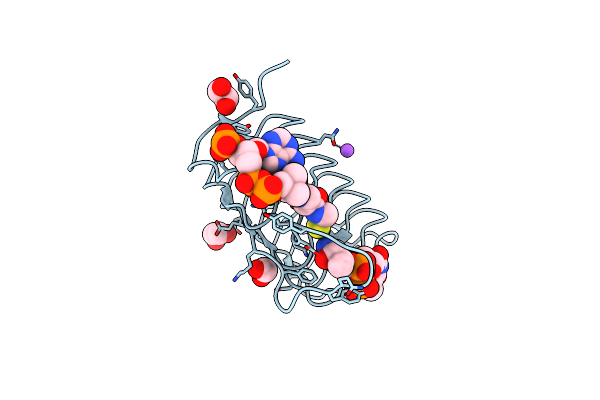 |
Crystal Structure Of An N-Acetyltransferase, C80T Mutant, From Helicobacter Pullorum In The Presence Of Coenzyme A And Dtdp-3-Amino-3,6-Dideoxy-D-Galactose
Organism: Helicobacter pullorum
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-09-22 Classification: TRANSFERASE Ligands: T3F, CAO, EDO, NA |
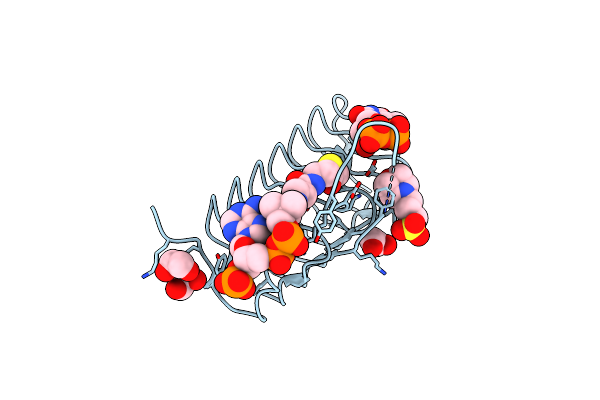 |
Crystal Structure Of An N-Acetyltransferase, C80T Mutant, From Helicobacter Pullorum In The Presence Of Acetyl Coenzyme A And Dtdp
Organism: Helicobacter pullorum
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2021-09-22 Classification: TRANSFERASE Ligands: TYD, ACO, EDO, MPO |
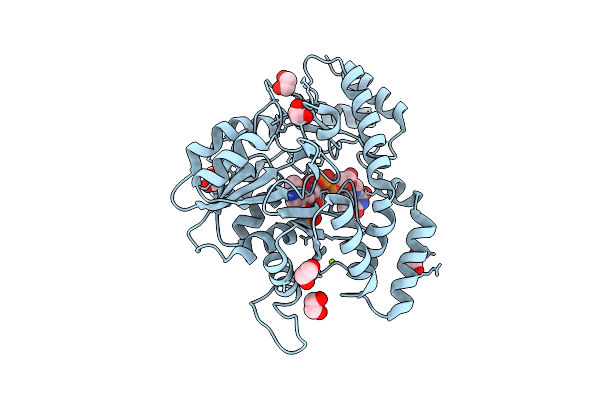 |
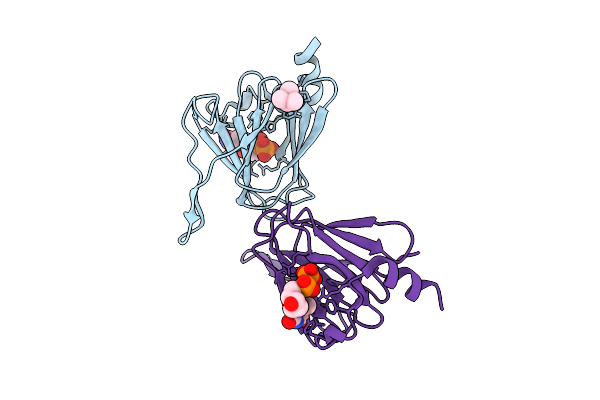
X-Ray Structure Of Hcan_0200, An Aminotransferase From Helicobacter Canadensis In Complex With Its External Aldimine
Organism: Helicobacter canadensis mit 98-5491
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-06-23 Classification: TRANSFERASE Ligands: TQP, MG, EDO, CL |
 |
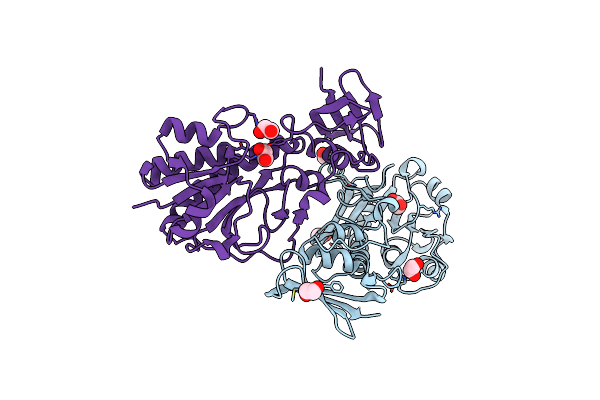
Crystal Structure Of Hcan_0198, A 3,4-Ketoisomerase From Helicobacter Canadensis
Organism: Helicobacter canadensis mit 98-5491
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-06-23 Classification: ISOMERASE Ligands: TYD, TMA |
 |
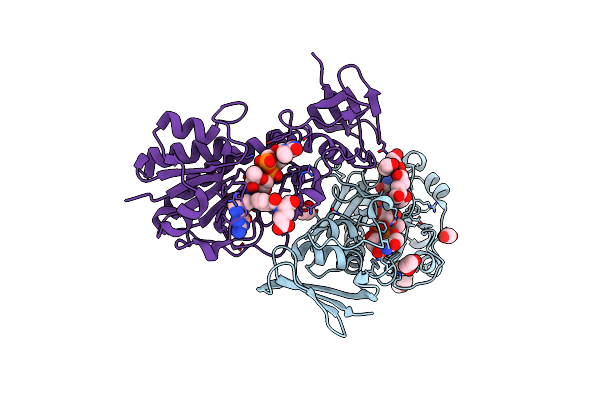
Crystal Structure Of The Dtdp-Qui3N N-Formyltransferase From Helicobacter Canadensis, Apo Form
Organism: Helicobacter canadensis mit 98-5491
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-06-23 Classification: TRANSFERASE Ligands: EDO |
 |
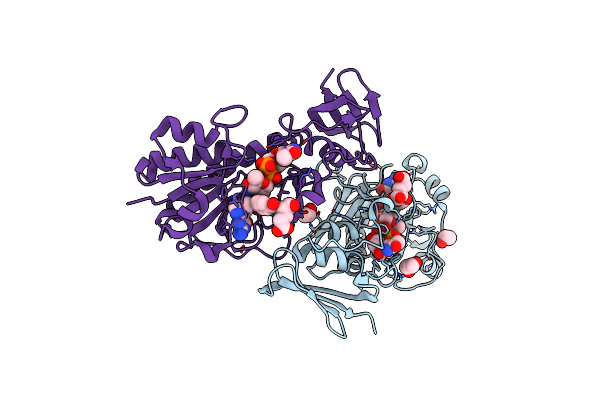
Crystal Structure Of The N-Formyltrasferase Hcan_0200 From Helicobacter Canadensis On Complex With Folinic Acid And Dtdp-3-Aminofucose
Organism: Helicobacter canadensis mit 98-5491
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-06-23 Classification: TRANSFERASE Ligands: FON, T3F, MES, EDO |
 |
Crystal Structure Of The N-Formyltransferase Hcan_0200 From Helicobacter Canadensis In Complex With Folinic Acid And Dtdp-3-Aminoquinovose
Organism: Helicobacter canadensis mit 98-5491
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-06-23 Classification: TRANSFERASE Ligands: FON, T3Q, EDO, K |

