Search Count: 66
 |
Structural Basis Of Specific Lysine Transport By Pseudomonas Aeruginosa Permease Lysp
Organism: Pseudomonas aeruginosa pao1, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MEMBRANE PROTEIN Ligands: LYS |
 |
Crystal Structure Of A Chimeric Alpha-Amylase From Pseudoalteromonas Haloplanktis Complexed With Rearranged Acarbose
Organism: Pseudoalteromonas haloplanktis
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2023-06-21 Classification: HYDROLASE Ligands: CL, CA, EDO |
 |
Crystal Structure Of A Chimeric Alpha-Amylase From Pseudoalteromonas Haloplanktis
Organism: Pseudoalteromonas haloplanktis
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2023-06-21 Classification: HYDROLASE Ligands: EDO, CL, CA |
 |
Crystal Structure Of A Second Homolog Of R2-Like Ligand-Binding Oxidase In Sulfolobus Acidocaldarius (Sar2Loxii)
Organism: Sulfolobus acidocaldarius dsm 639
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2022-04-06 Classification: OXIDOREDUCTASE Ligands: MN3, FE |
 |
Crystal Structure Of R2-Like Ligand-Binding Oxidase From Saccharopolyspora Erythraea
Organism: Saccharopolyspora erythraea (strain atcc 11635 / dsm 40517 / jcm 4748 / nbrc 13426 / ncimb 8594 / nrrl 2338)
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2022-04-06 Classification: OXIDOREDUCTASE Ligands: MN3, FE, PLM, SO4 |
 |
Organism: Saccharopolyspora erythraea (strain atcc 11635 / dsm 40517 / jcm 4748 / nbrc 13426 / ncimb 8594 / nrrl 2338)
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2021-03-03 Classification: TRANSCRIPTION Ligands: CO |
 |
Dtxr-Like Iron-Dependent Regulator Ider Complexed With Cobalt And Its Consensus Dna-Binding Sequence
Organism: Saccharopolyspora erythraea (strain atcc 11635 / dsm 40517 / jcm 4748 / nbrc 13426 / ncimb 8594 / nrrl 2338), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2021-03-03 Classification: TRANSCRIPTION Ligands: CO |
 |
Dtxr-Like Iron-Dependent Regulator Ider Complexed With Iron And Its Consensus Dna-Binding Sequence
Organism: Saccharopolyspora erythraea (strain atcc 11635 / dsm 40517 / jcm 4748 / nbrc 13426 / ncimb 8594 / nrrl 2338), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2021-03-03 Classification: TRANSCRIPTION Ligands: FE2 |
 |
Dtxr-Like Iron-Dependent Regulator Ider Complexed With Cobalt And The Sace_2689 Promoter Dna-Binding Sequence
Organism: Saccharopolyspora erythraea (strain atcc 11635 / dsm 40517 / jcm 4748 / nbrc 13426 / ncimb 8594 / nrrl 2338), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-03-03 Classification: TRANSCRIPTION Ligands: CO |
 |
Dtxr-Like Iron-Dependent Regulator Ider (P39G Variant) Complexed With Cobalt And Its Consensus Dna-Binding Sequence
Organism: Saccharopolyspora erythraea (strain atcc 11635 / dsm 40517 / jcm 4748 / nbrc 13426 / ncimb 8594 / nrrl 2338), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2021-03-03 Classification: TRANSCRIPTION Ligands: CO |
 |
Dtxr-Like Iron-Dependent Regulator Ider (Q43A Variant) Complexed With Cobalt And Its Consensus Dna-Binding Sequence
Organism: Saccharopolyspora erythraea (strain atcc 11635 / dsm 40517 / jcm 4748 / nbrc 13426 / ncimb 8594 / nrrl 2338), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2021-03-03 Classification: TRANSCRIPTION Ligands: CO |
 |
Crystal Structure Of Ribonucleotide Reductase Nrdf From Bacillus Anthracis Aerobically Soaked With Fe(Ii) And Mn(Ii) Ions
Organism: Bacillus anthracis str. sterne
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2020-04-15 Classification: OXIDOREDUCTASE Ligands: SO4, MN |
 |
Crystal Structure Of Ribonucleotide Reductase Nrdf From Bacillus Anthracis Anaerobically Soaked With Fe(Ii) And Mn(Ii) Ions
Organism: Bacillus anthracis str. sterne
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-04-15 Classification: OXIDOREDUCTASE Ligands: MN, SO4 |
 |
Crystal Structure Of Apo (Metal-Free) Ribonucleotide Reductase Nrdf L61G Variant From Bacillus Anthracis
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2020-04-15 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
Crystal Structure Of Ribonucleotide Reductase Nrdf L61G Variant From Bacillus Anthracis Anaerobically Soaked With Fe(Ii) And Mn(Ii) Ions
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2020-04-15 Classification: OXIDOREDUCTASE Ligands: MN, SO4 |
 |
Crystal Structure Of Ribonucleotide Reductase Nrdf L61G Variant From Bacillus Anthracis Aerobically Soaked With Fe(Ii) And Mn(Ii) Ions
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-04-15 Classification: OXIDOREDUCTASE Ligands: FE2, SO4, MN |
 |
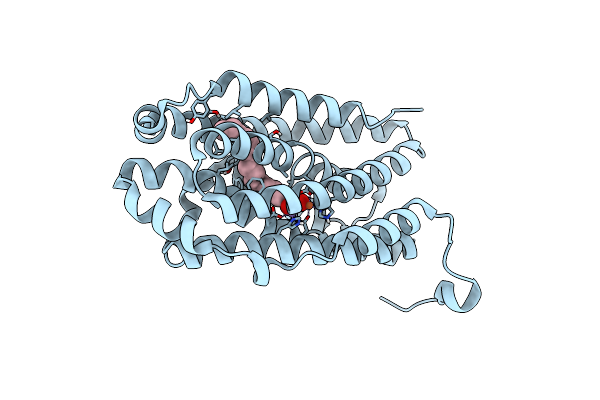
R2-Like Ligand-Binding Oxidase E69D Mutant With Anaerobically Reconstituted Mn/Fe Cofactor
Organism: Geobacillus kaustophilus (strain hta426)
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2020-02-19 Classification: OXIDOREDUCTASE Ligands: MN, FE2, PLM |
 |
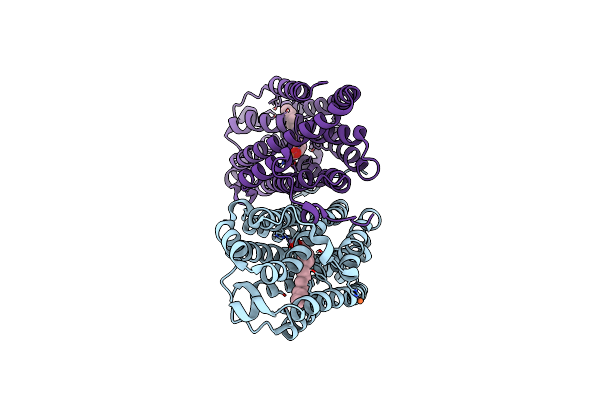
R2-Like Ligand-Binding Oxidase Y175F Mutant With Aerobically Reconstituted Mn/Fe Cofactor
Organism: Geobacillus kaustophilus hta426
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-02-19 Classification: OXIDOREDUCTASE Ligands: PLM, MN3, MN, FE |
 |
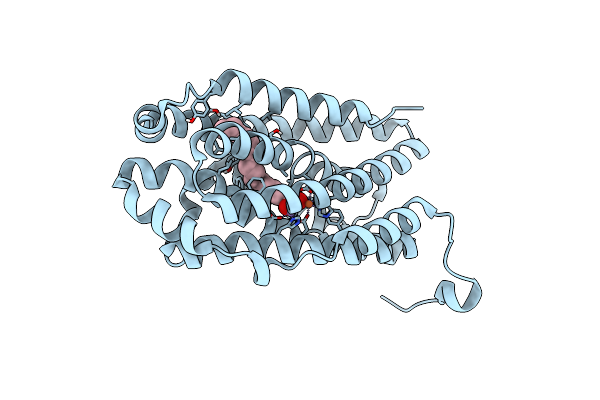
R2-Like Ligand-Binding Oxidase Y715F Mutant With Anaerobically Reconstituted Mn/Fe Cofactor
Organism: Geobacillus kaustophilus (strain hta426)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-02-19 Classification: OXIDOREDUCTASE Ligands: FE2, MN, PLM |
 |
R2-Like Ligand-Binding Oxidase E69D Mutant With Aerobically Reconstituted Mn/Fe Cofactor
Organism: Geobacillus kaustophilus (strain hta426)
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2020-02-12 Classification: OXIDOREDUCTASE Ligands: PLM, MN3, MN, FE |

