Search Count: 24
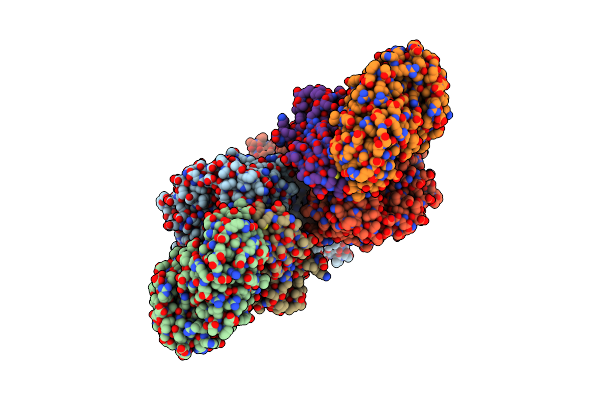 |
The Cryoem Structure Of The High Affinity Carbon Monoxide Dehydrogenase From Mycobacterium Smegmatis
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: CUN, MCN, FAD, FES |
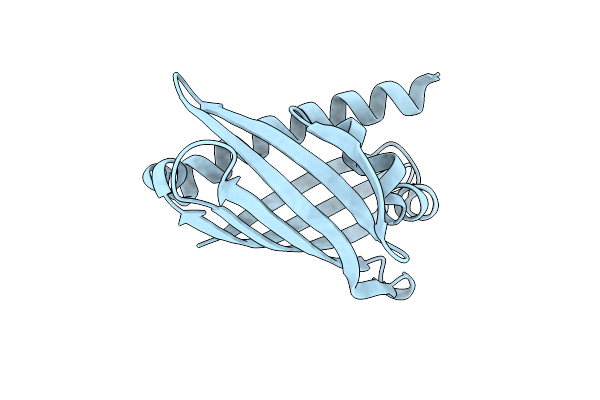 |
The Crystal Structure Of Coxg From M. Smegmatis, Minus Lipid Anchoring C-Terminus.
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Release Date: 2024-10-02 Classification: ELECTRON TRANSPORT |
 |
Organism: Bdellovibrio bacteriovorus hd100
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2023-10-25 Classification: UNKNOWN FUNCTION Ligands: GOL, EDO |
 |
Crystal Structure Of The N-Terminal Domain Of The Cryptic Surface Protein (Cd630_25440) From Clostridium Difficile.
Organism: Clostridioides difficile 630
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-05-10 Classification: UNKNOWN FUNCTION Ligands: CL, EDO, GOL |
 |
The 2.19-Angstrom Cryoem Structure Of The [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Complex Minus Stalk
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: 3NI, FCO, MG, MQ9, F3S |
 |
The 1.67 Angstrom Cryoem Structure Of The [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Catalytic Dimer (Huc2S2L)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: 3NI, FCO, OH, MG, VK3, F3S |
 |
The Cryoem Structure Of The [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Full Complex Focused Refinement Of Stalk
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: 3NI, FCO, MG, MQ9, F3S |
 |
The 1.52 Angstrom Cryoem Structure Of The [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Catalytic Dimer (Huc2S2L)
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2023-01-04 Classification: ELECTRON TRANSPORT Ligands: O, MG, FCO, 3NI, VK3, F3S |
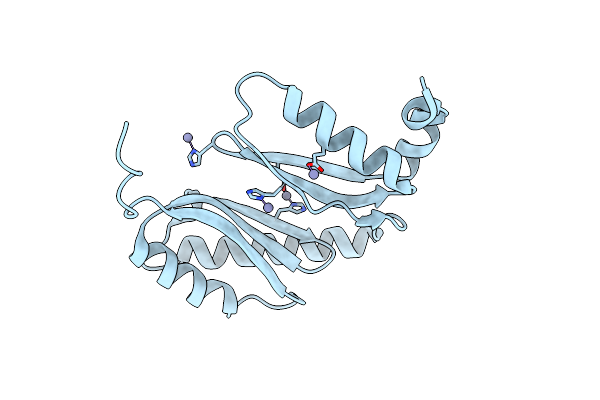 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-06-02 Classification: LIPID BINDING PROTEIN Ligands: ZN |
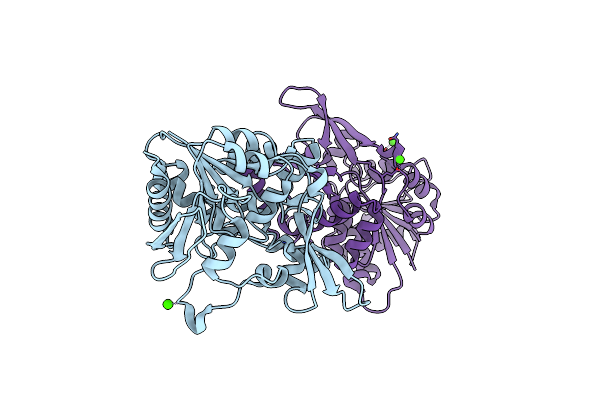 |
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-05-13 Classification: BIOSYNTHETIC PROTEIN Ligands: CA |
 |
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2020-05-13 Classification: BIOSYNTHETIC PROTEIN Ligands: FO1, CA |
 |
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-05-13 Classification: BIOSYNTHETIC PROTEIN Ligands: GDP, GOL, CA |
 |
The Crystal Structure Of Fbia From Mycobacterium Smegmatis, Gdp And Fo Bound Form
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-05-13 Classification: BIOSYNTHETIC PROTEIN Ligands: GDP, FO1, CA |
 |
The Crystal Structure Of Fbia From Mycobacterium Smegmatis, Dehydro-F420-0 Bound Form
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2020-05-13 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL, QMD, CA |
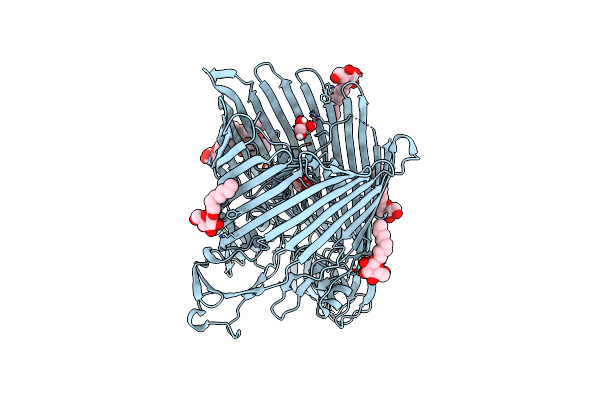 |
The Crystal Structure Of The Outer Membrane Transporter Yddb From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-10-02 Classification: TRANSPORT PROTEIN Ligands: BOG, MG, GOL |
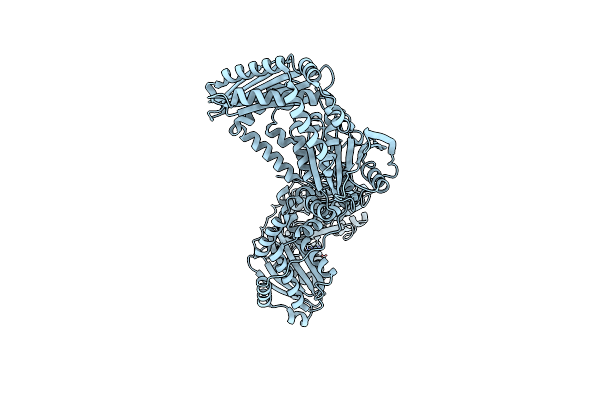 |
The Crystal Structure Of The Periplasmic Protease Pqql From Escherichia Coli
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-10-02 Classification: HYDROLASE Ligands: ZN, CL |
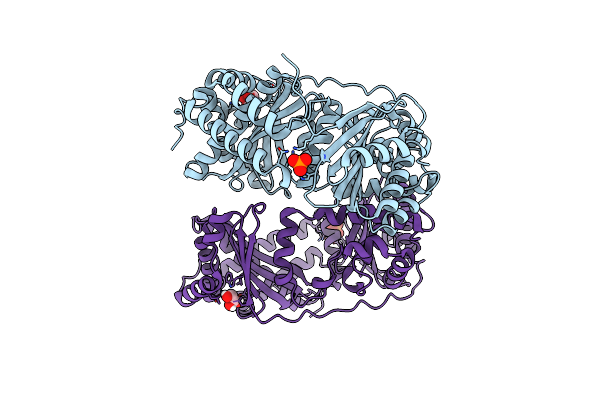 |
The Crystal Structure Of The First Half Of The Periplasmic Protease Pqql From Escherichia Coli
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-10-02 Classification: HYDROLASE Ligands: GOL, PO4, CL |
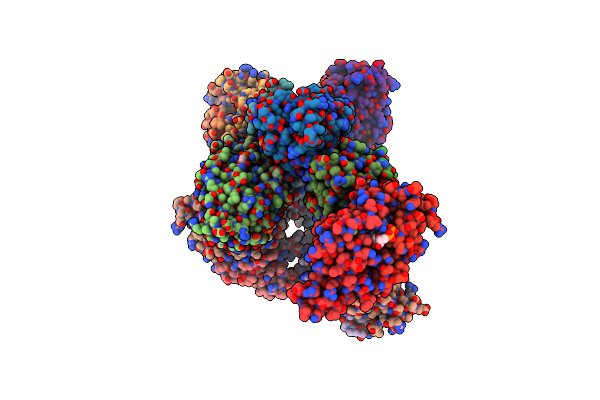 |
Structure Of The Fad Binding Protein Msmeg_5243 From Mycobacterium Smegmatis
Organism: Mycobacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2018-12-26 Classification: FAD-BINDING PROTEIN Ligands: FAD, CL |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2018-12-19 Classification: TRANSFERASE |
 |
Crystal Structure Of Mycoibacterium Tuberculosis Rv2983 In Complex With Pep
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2018-12-19 Classification: TRANSFERASE Ligands: PEP, MG |

