Search Count: 18
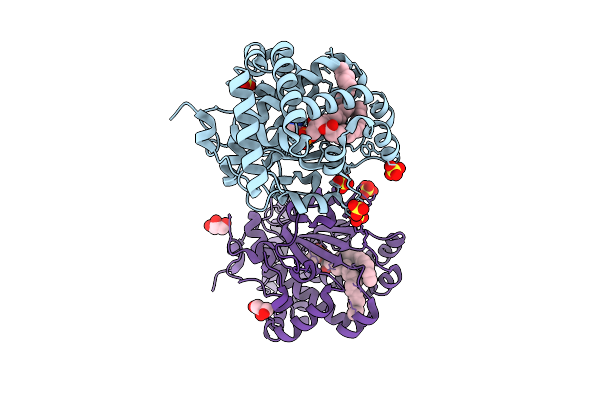 |
Crystal Structure Of Saccharomyces Cerevisiae Cross-Linked Sfh1 (K197C, F233C) Mutant In Complex With Phosphatidylethanolamine
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: LIPID BINDING PROTEIN Ligands: PTY, SO4, GOL |
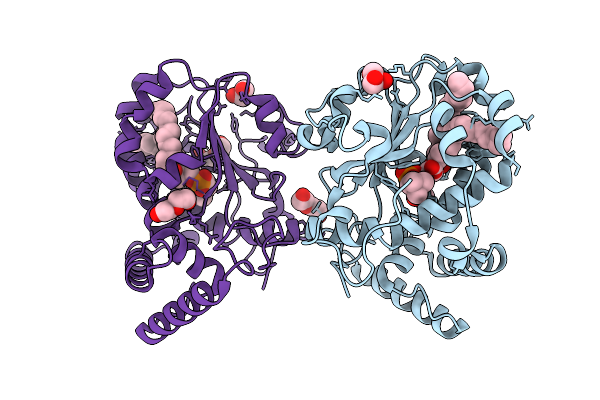 |
Crystal Structure Of Saccharomyces Cerevisiae Sec14P In Complex With Phosphatidylcholine
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: LIPID BINDING PROTEIN Ligands: POV, GOL, EDO, PEG |
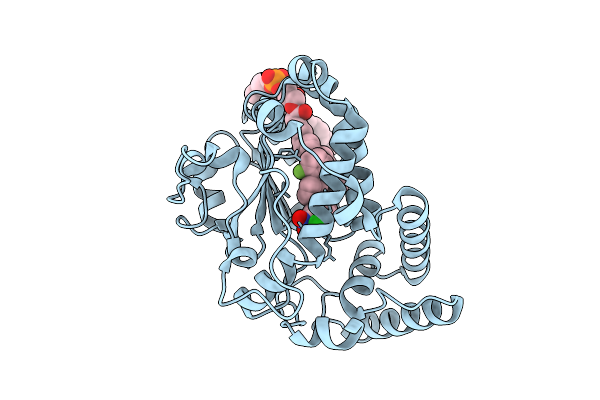 |
Crystal Structure Of A Ternary Complex Of Saccharomyces Cerevisiae Sec14 With A Small Molecule Inhibitor And Phosphatidylcholine
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: LIPID BINDING PROTEIN Ligands: POV, IUO |
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2023-01-25 Classification: LIPID BINDING PROTEIN Ligands: KO0 |
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-01-25 Classification: LIPID BINDING PROTEIN Ligands: IUF |
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-01-25 Classification: LIPID BINDING PROTEIN Ligands: IUJ |
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2023-01-25 Classification: LIPID BINDING PROTEIN Ligands: IUO, PO4 |
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2023-01-25 Classification: LIPID BINDING PROTEIN Ligands: IUC |
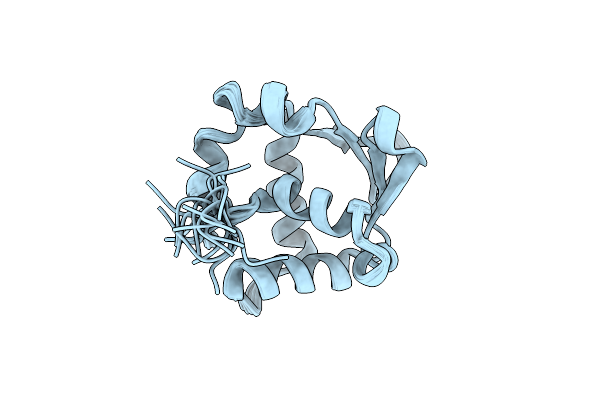 |
Intramolecular Regulation Of The Ets Domain Within Etv6 Sequence R335 To Q436
|
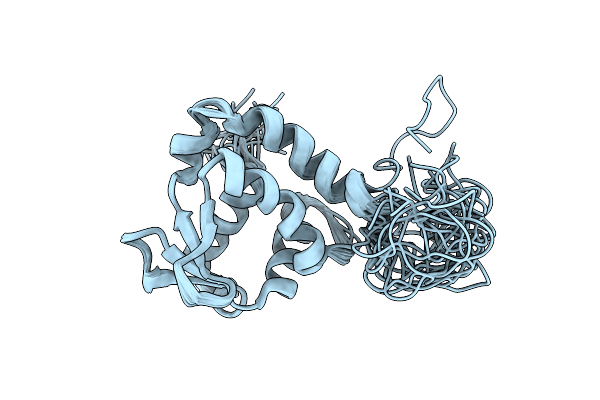 |
Intramolecular Regulation Of The Ets Domain Within Etv6 Sequence R335 To R458
|
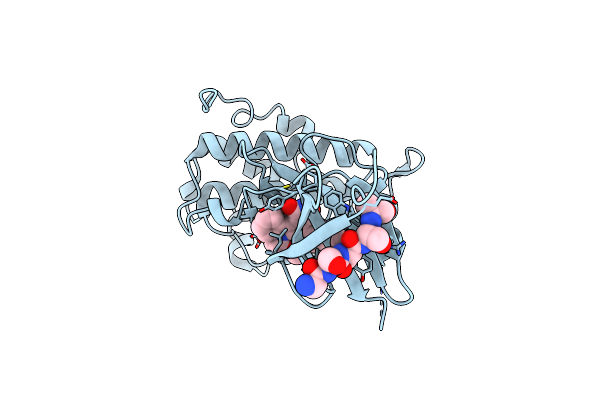 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-04-08 Classification: TRANSFERASE Ligands: SO4, UCN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2003-04-08 Classification: TRANSFERASE Ligands: SO4, STU |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2003-04-08 Classification: TRANSFERASE Ligands: SO4, UCM |
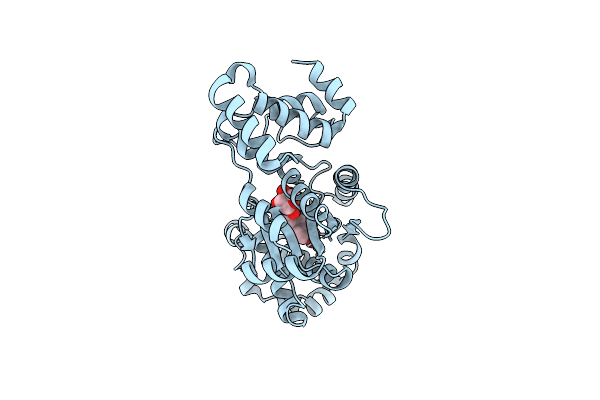 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2001-10-26 Classification: LIGASE Ligands: 383 |
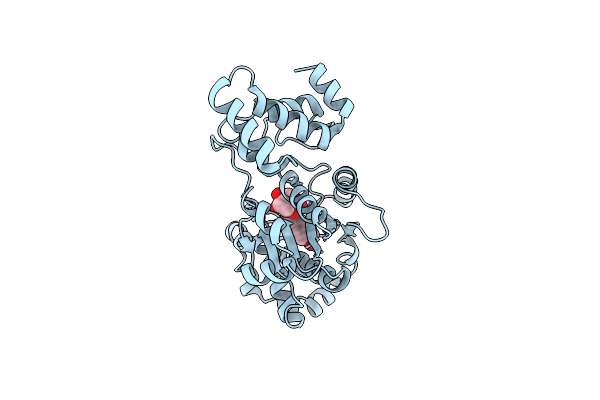 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2001-10-26 Classification: LIGASE Ligands: 629 |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2001-10-26 Classification: LIGASE Ligands: 545 |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2001-10-26 Classification: LIGASE Ligands: 485 |
 |
Staphylococcal Nuclease Dimer Containing A Deletion Of Residues 114-119 Complexed With Calcium Chloride And The Competitive Inhibitor Deoxythymidine-3',5'-Diphosphate
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 1997-04-21 Classification: HYDROLASE |

