Search Count: 78
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: A1BCZ, NA |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: A1BCY |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: A1BCX |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: A1BCW |
 |
Structure Of The G848S Mutant Of Human Mitochondrial Dna Polymerase Gamma In Complex With Pzl-A
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: TRANSFERASE/DNA Ligands: CA, DCP, A1IK1 |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: TRANSFERASE/DNA Ligands: CA, DCP |
 |
Structure Of The A467T Mutant Of Human Mitochondrial Dna Polymerase Gamma In Complex With Pzl-A
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: TRANSFERASE/DNA Ligands: CA, DCP, A1IK1 |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: TRANSFERASE/DNA Ligands: CA, DCP |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: TRANSFERASE/DNA Ligands: CA, DCP |
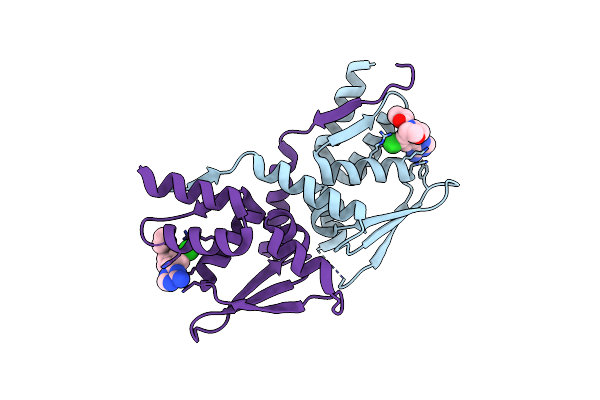 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2024-10-23 Classification: PROTEIN BINDING Ligands: A1BCT |
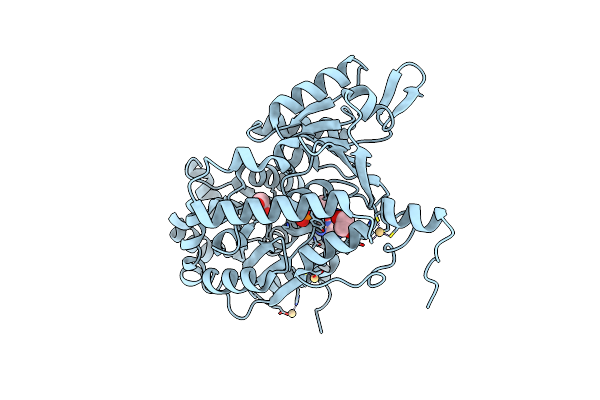 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2024-05-01 Classification: HYDROLASE Ligands: ADP, CD, EDO, CL |
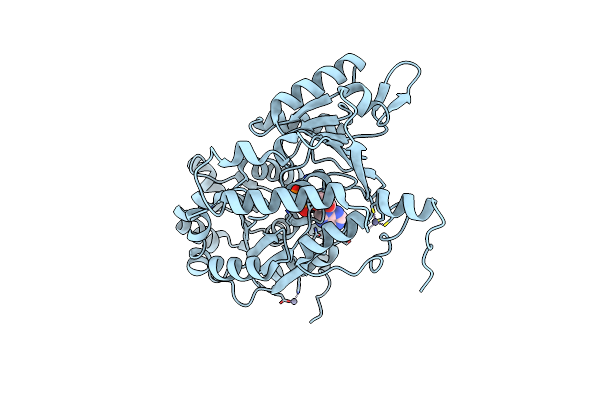 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2024-05-01 Classification: HYDROLASE Ligands: ZN, ATP, MG |
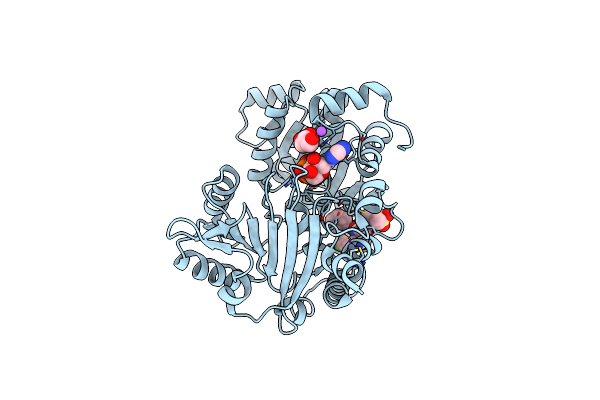 |
Crystal Structure Of Werner Helicase Fragment 517-945 In Covalent Complex With N-[(E,1S)-1-Cyclopropyl-3-Methylsulfonylprop-2-Enyl]-2-(1,1-Difluoroethyl)-4-Phenoxypyrimidine-5-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-05-01 Classification: HYDROLASE Ligands: ZN, ADP, X1L, GOL, NA |
 |
Organism: Aliivibrio fischeri
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-11-29 Classification: TRANSCRIPTION Ligands: SF4, MPD |
 |
Crystal Structure Of N-Myristoyl Transferase (Nmt) G386E Mutant From Plasmodium Vivax
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2019-06-05 Classification: TRANSFERASE Ligands: YNC, CL, EDO, SO4 |
 |
Crystal Structure Of N-Myristoyl Transferase (Nmt) G386E Mutant From Plasmodium Vivax In Complex With Inhibitor Imp-0366
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-06-05 Classification: TRANSFERASE Ligands: YNC, CL, 646, EDO, SO4 |
 |
Crystal Structure Of N-Myristoyl Transferase (Nmt) G386E Mutant From Plasmodium Vivax In Complex With Inhibitor Imp-1002
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-06-05 Classification: TRANSFERASE Ligands: YNC, JCY, SO4, CL, EDO |
 |
Crystal Structure Of N-Myristoyl Transferase (Nmt) From Plasmodium Vivax In Complex With Inhibitor Imp-1002
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Release Date: 2019-06-05 Classification: TRANSFERASE Ligands: YNC, JCY, CL, EDO, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2018-03-07 Classification: Transferase/Transferase Inhibitor Ligands: 8UV |
 |
Organism: Homo sapiens, Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-03-07 Classification: Transferase/Transferase Inhibitor Ligands: 504 |

