Search Count: 24
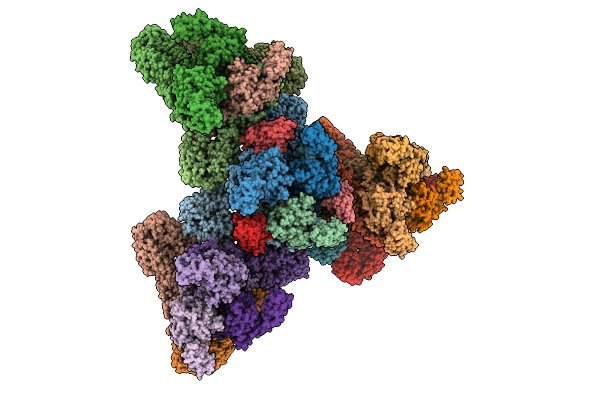 |
Venezuelan Equine Encephalitis Virus In Complex With The Single Domain Antibody V2B3
Organism: Venezuelan equine encephalitis virus, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: Viral protein/Immune System |
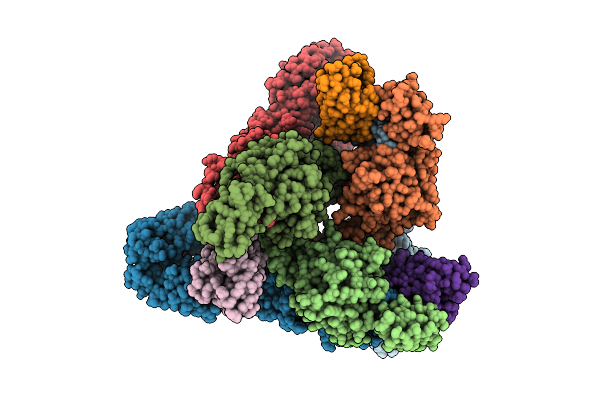 |
Venezuelan Equine Encephalitis Virus In Complex With The Single Domain Antibody V2C3
Organism: Venezuelan equine encephalitis virus, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: Viral protein/Immune System |
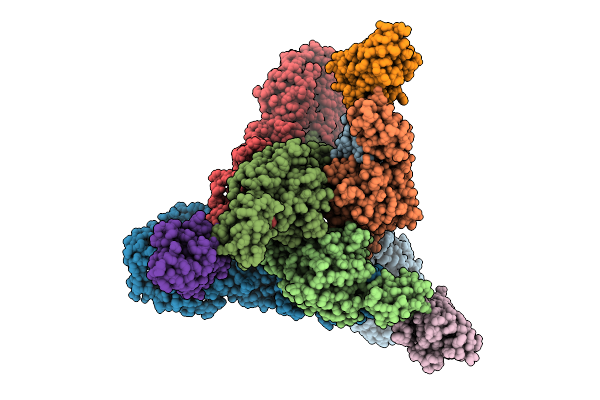 |
Venezuelan Equine Encephalitis Virus In Complex With The Single Domain Antibody V3A8F
Organism: Venezuelan equine encephalitis virus, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: Viral protein/Immune System |
 |
Organism: Erithacus rubecula
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: CIRCADIAN CLOCK PROTEIN |
 |
Crystal Structure Of Hla-B*15:01 In Complex With Spike Derived Peptide Nqklianaf From Oc43 Virus
Organism: Homo sapiens, Human coronavirus 229e
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-06-14 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: ACT |
 |
Crystal Structure Of Hla-B*15:01 In Complex With Spike Derived Peptide Nqklianqf From Sars-Cov-2 Virus
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-06-14 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: ACT, PEG |
 |
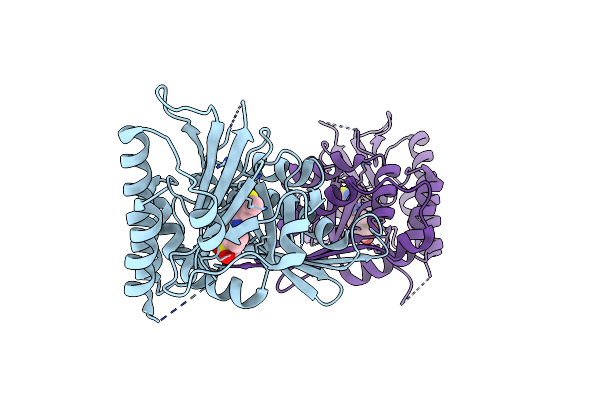
Crystal Structure Of Phosphatidylinositol 5-Phosphate 4-Kinase Type-2 Alpha (Pi5P4Ka) Bound To An Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-04-26 Classification: TRANSFERASE Ligands: U1U |
 |
Crystal Structure Of Phosphatidylinositol 5-Phosphate 4-Kinase (Pi5P4K2C) Bound To An Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2022-12-28 Classification: TRANSFERASE Ligands: QZR |
 |
Crystal Structure Of Phosphatidylinositol 5-Phosphate 4-Kinase (Pi5P4K2C) Bound To An Allosteric Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2022-05-18 Classification: TRANSFERASE Ligands: DVF |
 |
Crystal Structure Of Phosphatidylinositol 5-Phosphate 4-Kinase (Pi5P4K2C) Bound To An Allosteric Inhibitor And Amp-Pnp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-05-18 Classification: TRANSFERASE Ligands: DVF, ANP |
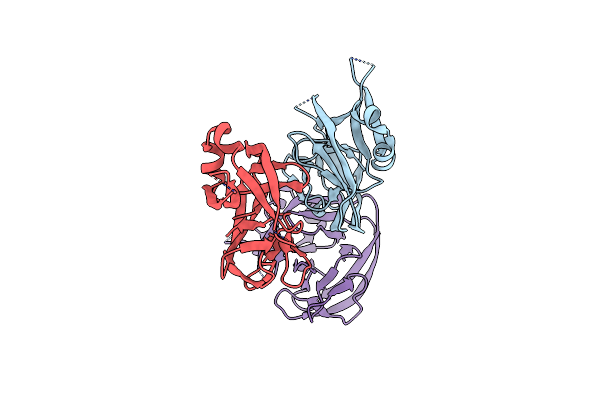 |
Organism: Cryptococcus gattii serotype b
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2019-11-06 Classification: HYDROLASE |
 |
Organism: Cryptococcus gattii serotype b
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2019-11-06 Classification: hydrolase/hydrolase inhibitor Ligands: PT |
 |
Organism: Cryptococcus neoformans var. grubii serotype a (strain h99 / atcc 208821 / cbs 10515 / fgsc 9487)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-10-23 Classification: HYDROLASE |
 |
Organism: Cryptococcus neoformans var. grubii
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2019-10-23 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of Pigeon Cryptochrome 4 Mutant Y319D In Complex With Flavin Adenine Dinucleotide
Organism: Columba livia
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2019-09-04 Classification: CIRCADIAN CLOCK PROTEIN Ligands: FAD, MG, PEG, GOL |
 |
Organism: Columba livia
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-09-04 Classification: CIRCADIAN CLOCK PROTEIN Ligands: FAD, PGE, EDO, PEG, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2017-01-25 Classification: DNA BINDING PROTEIN |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-09-21 Classification: SPLICING Ligands: CPT, TCE, SO4 |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2016-09-21 Classification: HYDROLASE Ligands: ZPT, SO4, TCE |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-10-01 Classification: CIRCADIAN CLOCK PROTEIN/TRANSCRIPTION Ligands: ZN |

