Search Count: 70
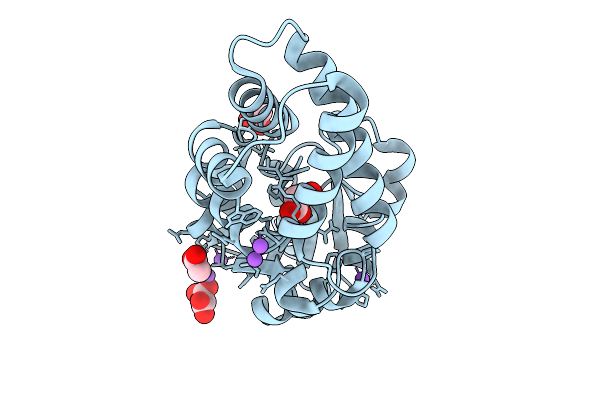 |
Crystal Structure Of An Engineered Enzyme Containing A Genetically Encoded Thiourea, Ct1.5
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: BIOSYNTHETIC PROTEIN Ligands: MLA, ACY, NA |
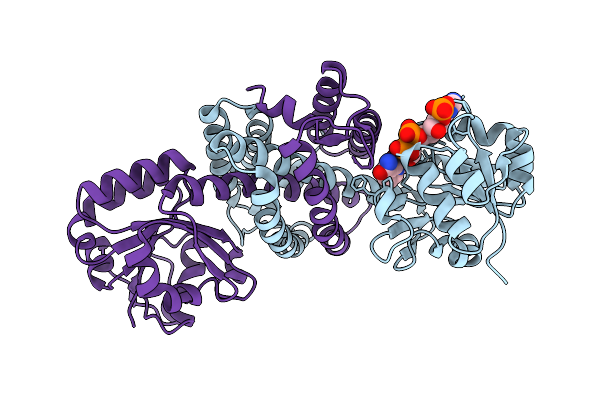 |
Structure Of Imine Reductase 361 From Micromonospora Sp. Mutant M125W/I127F/L179V/H250L
Organism: Micromonospora
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: OXIDOREDUCTASE Ligands: NAP |
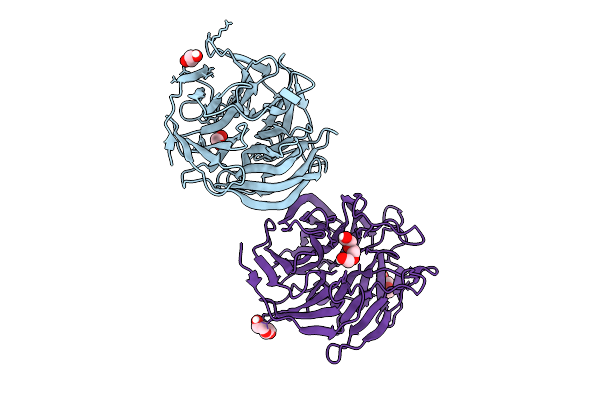 |
Crystal Structure Of Complement1.4 (Cent1.4), An Engineered Photoenzyme For Selective [2+2]-Cycloadditions
Organism: Loligo vulgaris
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO, PEG, PGE |
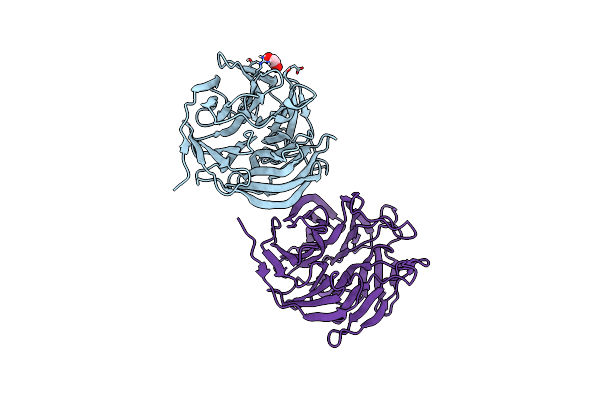 |
Organism: Loligo vulgaris
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO |
 |
Organism: Loligo vulgaris
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: BIOSYNTHETIC PROTEIN Ligands: PEG, EDO |
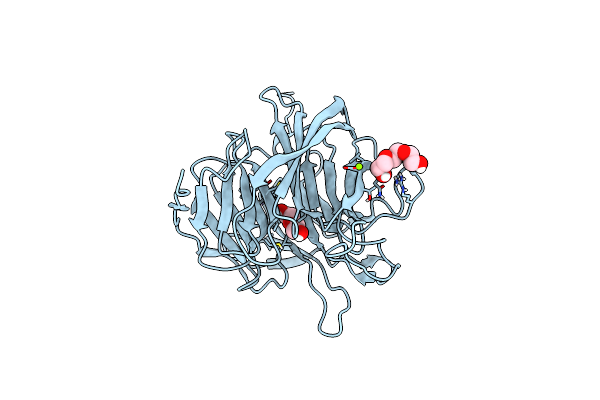 |
Organism: Loligo vulgaris
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: BIOSYNTHETIC PROTEIN Ligands: PEG, PGE, MG |
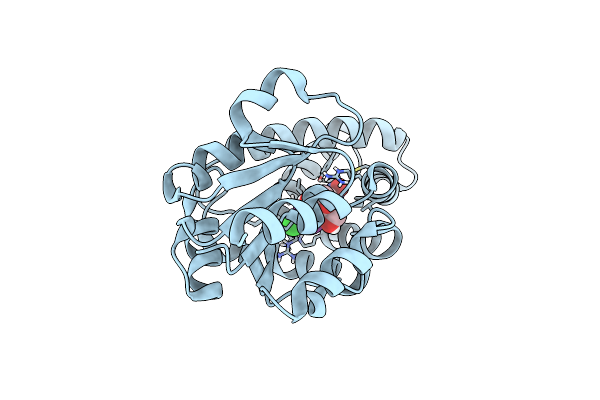 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2025-01-29 Classification: BIOSYNTHETIC PROTEIN Ligands: PEG, A1IGD, CL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2025-01-29 Classification: BIOSYNTHETIC PROTEIN Ligands: IOD, PG4 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2025-01-29 Classification: BIOSYNTHETIC PROTEIN Ligands: PG4, PEG, CL, PGE |
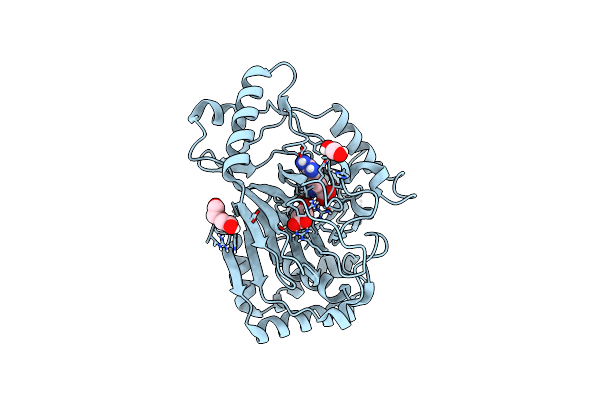 |
Crystal Structure Of The L-Arginine Hydroxylase Vioc Mehis316, Bound To Fe(Ii), L-Arginine, And Succinate
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-07-31 Classification: OXIDOREDUCTASE Ligands: FE2, SIN, ARG, EDO, PEG |
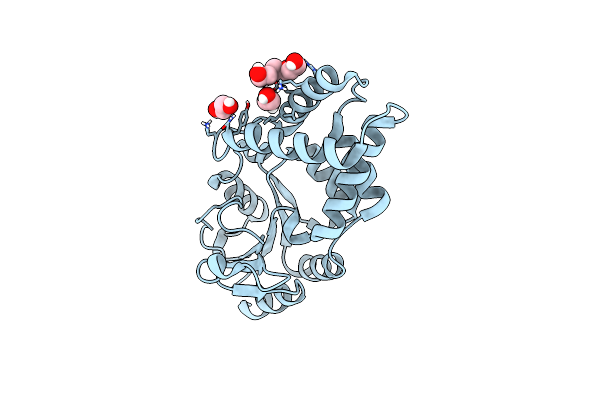 |
Crystal Structure Of Bhmehis1.8, An Engineered Enzyme For The Morita-Baylis-Hillman Reaction
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2024-02-28 Classification: BIOSYNTHETIC PROTEIN Ligands: PGE, EDO |
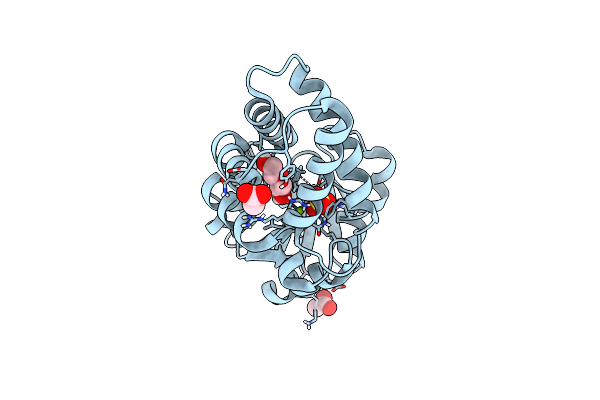 |
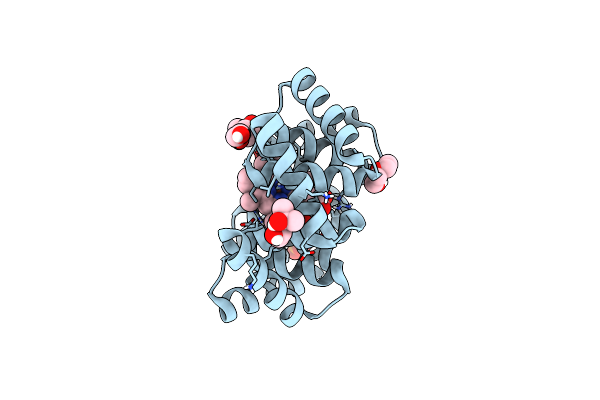
Crystal Structure Of Bhmehis1.0, An Engineered Enzyme For The Morita-Baylis-Hillman Reaction
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2024-02-28 Classification: BIOSYNTHETIC PROTEIN Ligands: PEG, ACT, MG, SO4 |
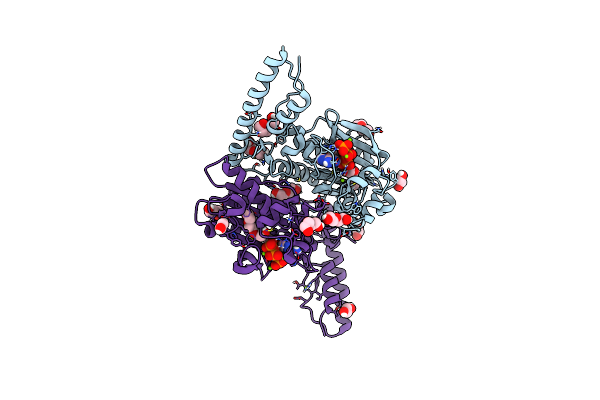 |
Crystal Structure Of Pyrrolysyl-Trna Synthetase From Methanomethylophilus Alvus Engineered For 3-Methyl-L-Histidine, Bound To Amppnp
Organism: Candidatus methanomethylophilus alvus
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2023-07-19 Classification: LIGASE Ligands: ANP, PEG, PGE, EDO, MG |
 |
Crystal Structure Of A Computationally Designed Heme Binding Protein, Dnhem1
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-07-05 Classification: BIOSYNTHETIC PROTEIN Ligands: IMD, HEM, MPD, PO4, PEG, EDO |
 |
Organism: Acinetobacter baumannii ab0057
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: Ribosome/RNA Ligands: ZN, OI9 |
 |
Organism: Acinetobacter baumannii ab0057
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: Ribosome/RNA Ligands: ZN |
 |
Organism: Acinetobacter baumannii ab0057
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: Ribosome/RNA Ligands: ZN, OI9 |
 |
Organism: Acinetobacter baumannii ab0057
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: Ribosome/RNA Ligands: ZN, OIY, MG |
 |
Organism: Acinetobacter baumannii ab0057
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: Ribosome/RNA Ligands: ZN, OIY, MG |
 |
Organism: Acinetobacter baumannii ab0057
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: Ribosome/RNA Ligands: ZN, OIY, MG |

