Search Count: 69
 |
Organism: Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2020-06-03 Classification: MEMBRANE PROTEIN Ligands: CPL, PX2, NA |
 |
Organism: Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2020-06-03 Classification: MEMBRANE PROTEIN Ligands: PCG, CPL, PX2 |
 |
Structure Of Cgmp-Unbound F403V/V407A Mutant Tax-4 Reconstituted In Lipid Nanodiscs
Organism: Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2020-06-03 Classification: MEMBRANE PROTEIN Ligands: CPL, PX2, NA |
 |
Organism: Escherichia coli, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2019-06-26 Classification: RIBOSOME Ligands: MG, ZN |
 |
Rf1 Accommodated State Bound Release Complex 70S At Long Incubation Time Point
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:3.50 Å Release Date: 2019-06-26 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Escherichia coli, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2019-06-19 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Escherichia coli, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2019-06-19 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Escherichia coli, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2019-06-19 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Escherichia coli, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2019-06-19 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Escherichia coli, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2019-06-19 Classification: RIBOSOME Ligands: MG, ZN |
 |
Cryo-Em Structure Of Lipid Bilayer In The Native Cell Membrane Nanoparticles Of Acrb
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2018-12-19 Classification: MEMBRANE PROTEIN Ligands: PTY, D12 |
 |
Single Particles Cryo-Em Structure Of Acrb D407A Associated With Lipid Bilayer At 3.0 Angstrom
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2018-12-05 Classification: TRANSPORT PROTEIN Ligands: PTY, D12 |
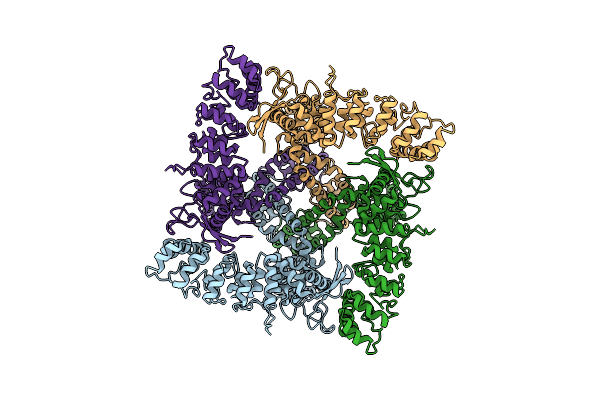 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2017-12-20 Classification: TRANSPORT PROTEIN |
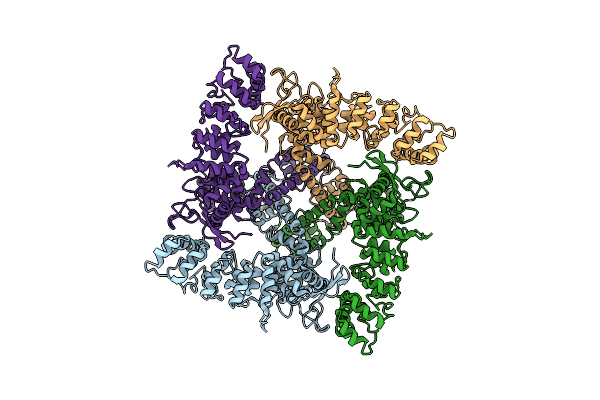 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2017-12-20 Classification: TRANSPORT PROTEIN |
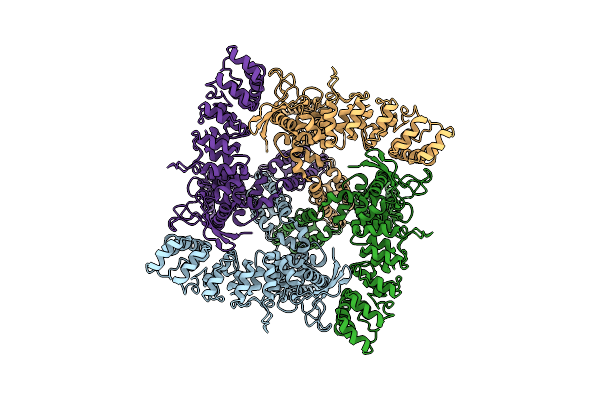 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2017-12-20 Classification: TRANSPORT PROTEIN |
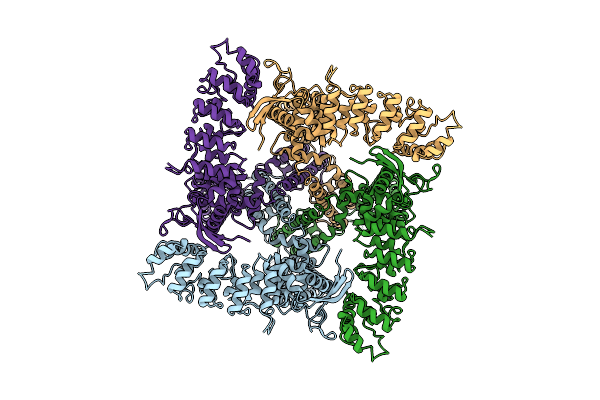 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2017-12-20 Classification: TRANSPORT PROTEIN |
 |
Organism: Rattus norvegicus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2017-08-02 Classification: TRANSPORT PROTEIN Ligands: ZK1 |
 |
Organism: Rattus norvegicus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2017-08-02 Classification: TRANSPORT PROTEIN Ligands: ZK1, AJP |
 |
Organism: Rattus norvegicus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2017-08-02 Classification: TRANSPORT PROTEIN |
 |
Organism: Rattus norvegicus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2017-08-02 Classification: TRANSPORT PROTEIN Ligands: AJP |

