Search Count: 57
 |
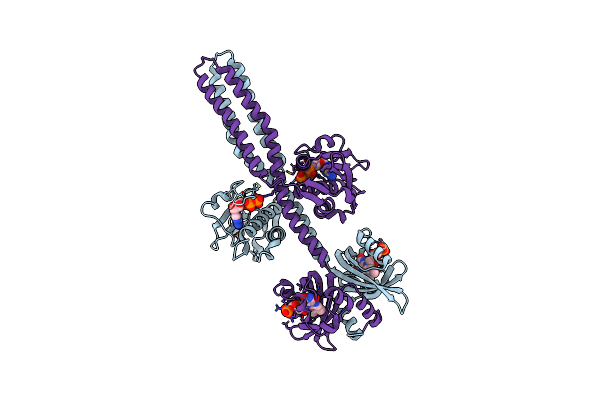
Crystal Structure Of A Chimeric Lov-Histidine Kinase Sb2F1 (Asymmetrical Variant, Trigonal Form With Long C Axis)
Organism: Pseudomonas putida kt2440, Bradyrhizobium diazoefficiens usda 110
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2024-01-10 Classification: SIGNALING PROTEIN Ligands: ATP, FMN |
 |
Crystal Structure Of A Chimeric Lov-Histidine Kinase Sb2F1-I66R Mutant (Asymmetrical Variant, Trigonal Form With Long C Axis)
Organism: Pseudomonas putida kt2440, Bradyrhizobium diazoefficiens (strain jcm 10833 / bcrc 13528 / iam 13628 / nbrc 14792 / usda 110)
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2024-01-10 Classification: SIGNALING PROTEIN Ligands: ATP, FMN |
 |
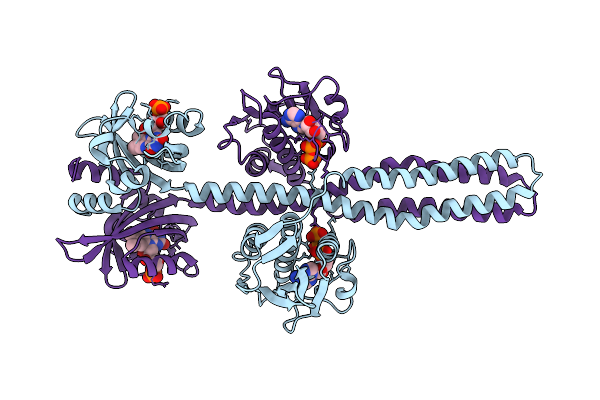
Crystal Structure Of A Chimeric Lov-Histidine Kinase Sb2F1-I66R Mutant (Light State; Asymmetrical Variant, Trigonal Form With Long C Axis)
Organism: Pseudomonas putida kt2440, Bradyrhizobium diazoefficiens (strain jcm 10833 / bcrc 13528 / iam 13628 / nbrc 14792 / usda 110)
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2024-01-10 Classification: SIGNALING PROTEIN Ligands: ATP, JGC |
 |
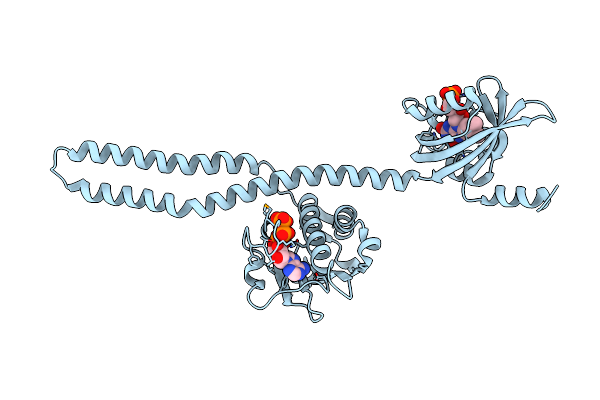
Crystal Structure Of A Chimeric Lov-Histidine Kinase Sb2F1 (Asymmetrical Variant, Trigonal Form With Long C-Axis)
Organism: Pseudomonas putida kt2440, Bradyrhizobium diazoefficiens usda 110
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2023-12-27 Classification: SIGNALING PROTEIN Ligands: ATP, FMN |
 |
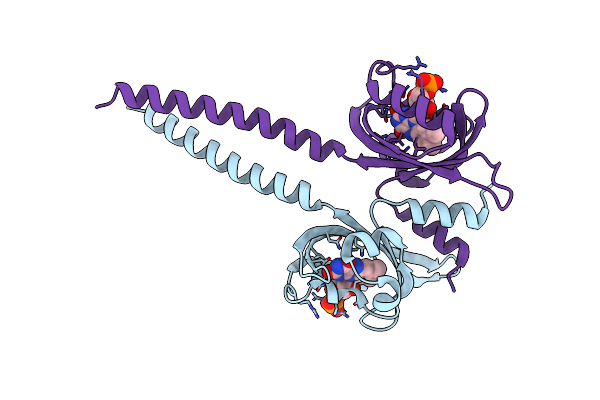
Crystal Structure Of A Chimeric Lov-Histidine Kinase Sb2F1 (Symmetrical Variant, Trigonal Form With Short C-Axis)
Organism: Pseudomonas putida kt2440, Bradyrhizobium diazoefficiens usda 110
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2023-12-20 Classification: SIGNALING PROTEIN Ligands: ATP, FMN |
 |
Crystal Structure Of The Full-Length Short Lov Protein Pf5-Lov From Pseudomonas Fluorescens (Dark State)
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2023-08-23 Classification: SIGNALING PROTEIN Ligands: FMN |
 |
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2023-07-05 Classification: SIGNALING PROTEIN Ligands: JGC |
 |
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-07-05 Classification: SIGNALING PROTEIN Ligands: FMN |
 |
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2023-07-05 Classification: SIGNALING PROTEIN Ligands: FMN, PEG |
 |
Crystal Structure Of The Full-Length Short Lov Protein Sbw25-Lov From Pseudomonas Fluorescens (Light State)
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Release Date: 2023-05-24 Classification: SIGNALING PROTEIN Ligands: FMN, JGC, ACT, NA |
 |
A Fast Recovering Full-Length Lov Protein (Dslov) From The Marine Phototrophic Bacterium Dinoroseobacter Shibae (Dark State) - C72A Mutant
Organism: Dinoroseobacter shibae dfl 12 = dsm 16493
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-05-17 Classification: SIGNALING PROTEIN Ligands: FMN |
 |
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2022-12-21 Classification: SIGNALING PROTEIN Ligands: FMN |
 |
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2022-12-21 Classification: SIGNALING PROTEIN Ligands: FMN |
 |
Structure Of Chloroflexus Aggregans Flavin Based Fluorescent Protein (Cagfbfp) Q148K Variant With Morpholine
Organism: Chloroflexus aggregans (strain md-66 / dsm 9485)
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2021-04-21 Classification: FLUORESCENT PROTEIN Ligands: FMN, 6LR, GOL |
 |
Structure Of Chloroflexus Aggregans Flavin Based Fluorescent Protein (Cagfbfp) Q148K Variant (No Morpholine)
Organism: Chloroflexus aggregans (strain md-66 / dsm 9485)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-04-21 Classification: FLUORESCENT PROTEIN Ligands: FMN, SO4 |
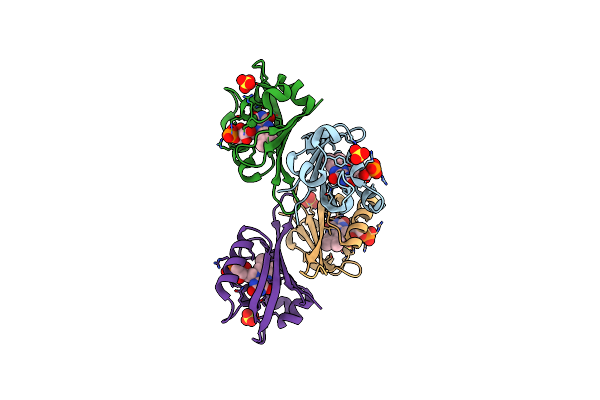 |
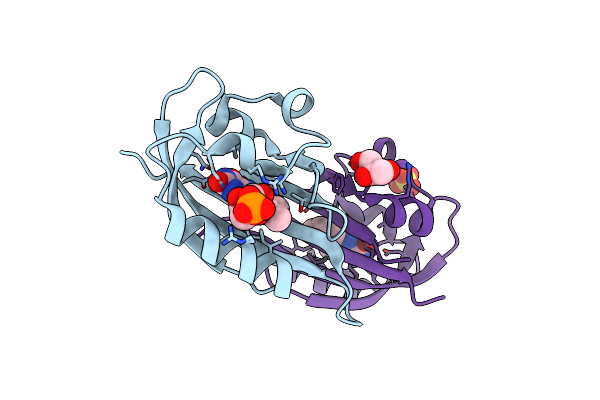
Structure Of Chloroflexus Aggregans Flavin Based Fluorescent Protein (Cagfbfp) Q148K Variant (Space Group P21)
Organism: Chloroflexus aggregans (strain md-66 / dsm 9485)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-04-21 Classification: FLUORESCENT PROTEIN Ligands: FMN, SO4 |
 |
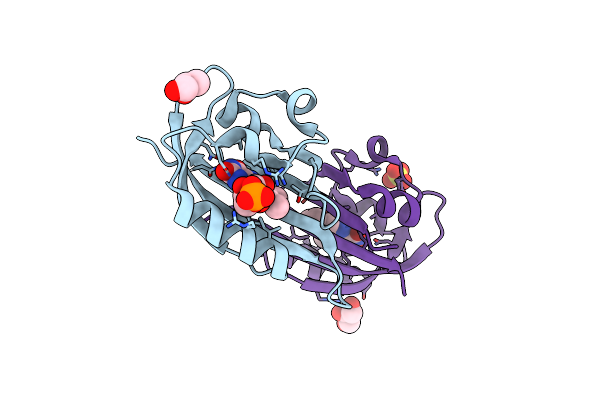
Structure Of Chloroflexus Aggregans Flavin Based Fluorescent Protein (Cagfbfp) I52T Variant
Organism: Chloroflexus aggregans (strain md-66 / dsm 9485)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-04-21 Classification: FLUORESCENT PROTEIN Ligands: FMN, GOL, SO4 |
 |
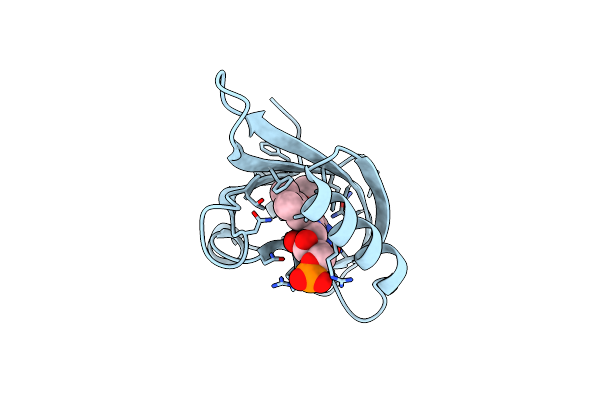
Structure Of Chloroflexus Aggregans Flavin Based Fluorescent Protein (Cagfbfp) I52T Q148K Variant
Organism: Chloroflexus aggregans (strain md-66 / dsm 9485)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-04-21 Classification: FLUORESCENT PROTEIN Ligands: FMN, GOL, SO4 |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-04-21 Classification: SIGNALING PROTEIN Ligands: FMN, ACT |
 |
Structural Determinants Underlying The Adduct Lifetime In A Short Lov Protein Ppsb2-Lov
Organism: Pseudomonas putida (strain atcc 47054 / dsm 6125 / ncimb 11950 / kt2440)
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2021-03-10 Classification: SIGNALING PROTEIN Ligands: PEG, FMN |

