Search Count: 37
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-06-28 Classification: OXIDOREDUCTASE Ligands: FAD, ACT |
 |
Phycocyanin Structure From A Modular Droplet Injector For Serial Femtosecond Crystallography
Organism: Thermosynechococcus vestitus bp-1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-06-28 Classification: PLANT PROTEIN Ligands: CYC, NA |
 |
Transitional Unit Cell 1 Of Adenine Riboswitch Aptamer Crystal Phase Transition Upon Ligand Binding
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2020-11-11 Classification: RNA Ligands: ADE, MG |
 |
Transitional Unit Cell 2 Of Adenine Riboswitch Aptamer Crystal Phase Transition Upon Ligand Binding
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-11-11 Classification: RNA Ligands: ADE, K, MG |
 |
Room Temperature Structure Of Nsp15 Endoribonuclease From Sars Cov-2 Solved Using Sfx.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-10-21 Classification: VIRAL PROTEIN Ligands: CIT |
 |
Organism: Escherichia coli (strain b / bl21-de3)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-09-02 Classification: TRANSFERASE |
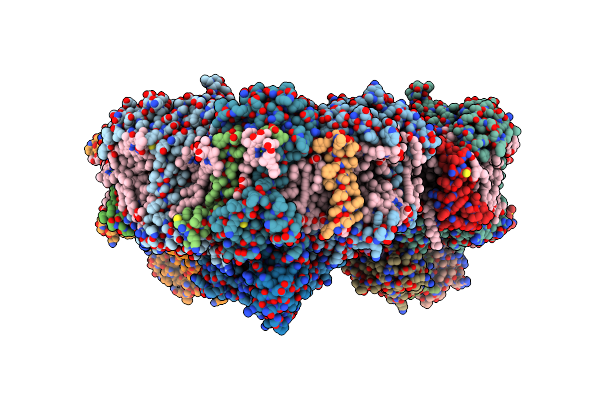 |
Membrane Protein Megahertz Crystallography At The European Xfel, Photosystem I Xfel At 2.9 A
Organism: Thermosynechococcus elongatus (strain bp-1)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2019-11-27 Classification: PHOTOSYNTHESIS Ligands: CL0, CLA, PQN, SF4, BCR, LHG, LMG, CA |
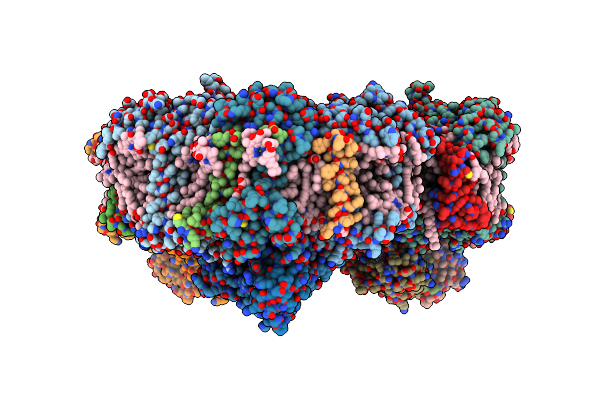 |
Membrane Protein Megahertz Crystallography At The European Xfel, Photosystem I At Synchrotron To 2.9 A
Organism: Thermosynechococcus elongatus (strain bp-1)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2019-11-20 Classification: PHOTOSYNTHESIS Ligands: CL0, CLA, PQN, SF4, BCR, LHG, LMG, CA |
 |
Xfel Crystal Structure Of Human Melatonin Receptor Mt1 In Complex With Ramelteon
Organism: Homo sapiens, Pyrococcus abyssi ge5
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-04-24 Classification: MEMBRANE PROTEIN Ligands: JEV, OLA, PEG |
 |
Xfel Crystal Structure Of Human Melatonin Receptor Mt1 In Complex With 2-Phenylmelatonin
Organism: Homo sapiens, Pyrococcus abyssi ge5
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2019-04-24 Classification: MEMBRANE PROTEIN Ligands: JEY, OLA, PEG |
 |
Xfel Crystal Structure Of Human Melatonin Receptor Mt1 In Complex With 2-Iodomelatonin
Organism: Homo sapiens, Pyrococcus abyssi ge5
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2019-04-24 Classification: MEMBRANE PROTEIN Ligands: ML2, PEG, GOL |
 |
Xfel Crystal Structure Of Human Melatonin Receptor Mt1 In Complex With Agomelatine
Organism: Homo sapiens, Pyrococcus abyssi ge5
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2019-04-24 Classification: MEMBRANE PROTEIN Ligands: AWY, OLA |
 |
Cs-Rosetta Determined Structures Of The N-Terminal Domain Of Algf From P. Aeruginosa
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: SOLUTION NMR Release Date: 2019-04-17 Classification: CELL ADHESION |
 |
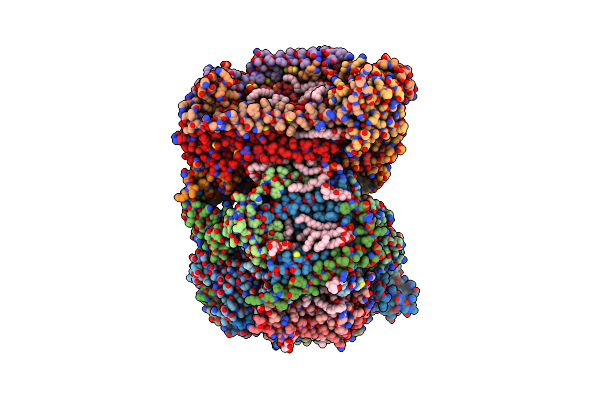
Cs-Rosetta Determined Structures Of The C-Terminal Domain Of Algf From P. Aeruginosa
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: SOLUTION NMR, SOLUTION SCATTERING Release Date: 2019-04-17 Classification: CELL ADHESION |
 |
Time-Resolved Sfx Structure Of The Pr Intermediate Of Cytochrome C Oxidase At Room Temperature
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-03-20 Classification: OXIDOREDUCTASE Ligands: CU, MG, NA, HEA, PGV, TGL, O, OH, CUA, PSC, CHD, PEK, CDL, ZN, DMU |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-03-13 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, PGV, TGL, CDL, CUA, CHD, PSC, DMU, PEK, ZN |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2019-03-13 Classification: OXIDOREDUCTASE Ligands: CU, MG, NA, HEA, PGV, TGL, PER, CUA, PSC, CHD, DMU, PEK, CDL, ZN |
 |
Crystal Structure Of Co-Bound Cytochrome C Oxidase Determined By Serial Femtosecond X-Ray Crystallography At Room Temperature
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-08-09 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, PGV, TGL, CMO, CUA, PSC, CHD, PEK, CDL, DMU, ZN |
 |
Crystal Structure Of Co-Bound Cytochrome C Oxidase Determined By Synchrotron X-Ray Crystallography At 100 K
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-08-09 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, PGV, TGL, CMO, CUA, CHD, PEK, CDL, DMU, PSC, ZN |
 |
Organism: Homo sapiens, Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2017-05-24 Classification: SIGNALING PROTEIN Ligands: NAG, 97V |

