Search Count: 34
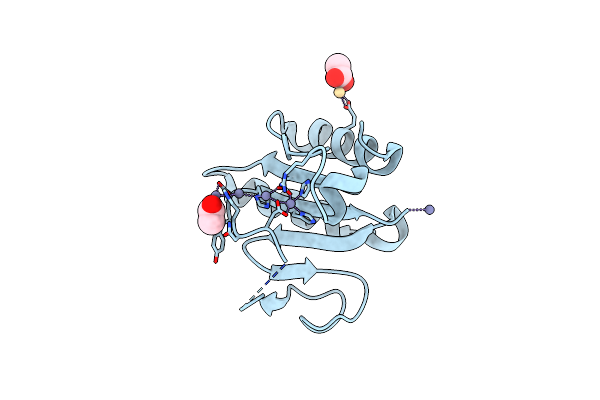 |
Crystal Structure Of The C-Terminal Domain Of Vlde From Streptococcus Pneumoniae Containing Four Zinc Atoms At The Binding Site
Organism: Streptococcus pneumoniae r6
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-01-22 Classification: METAL BINDING PROTEIN Ligands: ZN, CD, ACT |
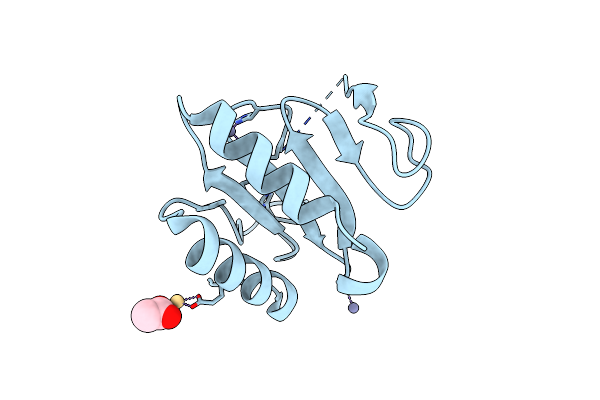 |
Crystal Structure Of The C-Terminal Domain Of Vlde From Streptococcus Pneumoniae Containing Three Zinc Atoms At The Binding Site
Organism: Streptococcus pneumoniae r6
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-01-22 Classification: METAL BINDING PROTEIN Ligands: ACT, ZN, CD |
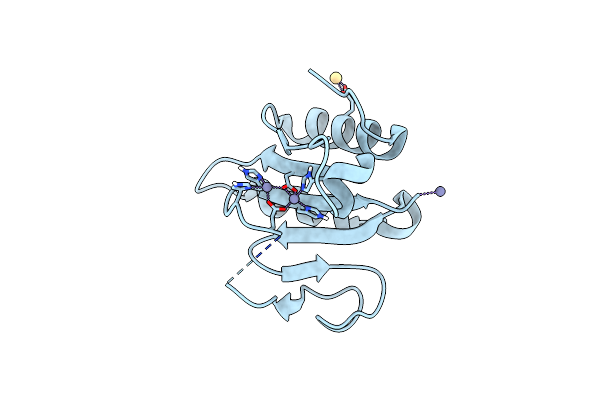 |
Crystal Structure Of The C-Terminal Domain Of Vlde From Streptococcus Pneumoniae Containing Two Zinc Atoms At The Binding Site
Organism: Streptococcus pneumoniae r6
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-01-22 Classification: METAL BINDING PROTEIN Ligands: ZN, CD |
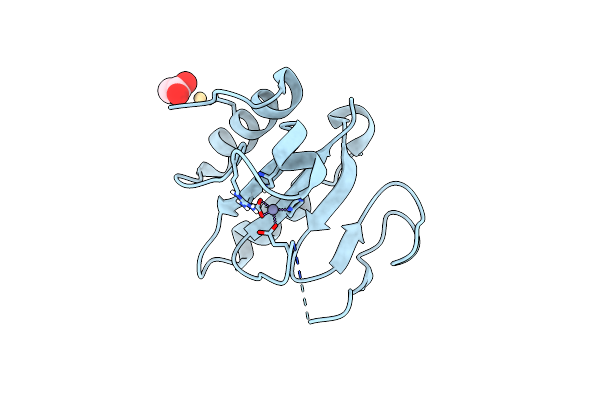 |
Crystal Structure Of The C-Terminal Domain Of Vlde From Streptococcus Pneumoniae Containing A Zinc Atom At The Binding Site
Organism: Streptococcus pneumoniae r6
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-01-22 Classification: METAL BINDING PROTEIN Ligands: ACT, CD, ZN |
 |
Crystal Structure Of The C-Terminal Domain Of Vlde From Streptococcus Pneumoniae In A Catalytically Competent Conformation
Organism: Streptococcus pneumoniae r6
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-01-22 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Crystal Structure Of The C-Terminal Domain Of Vlde H373A From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae r6
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2025-01-22 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Organism: Lactiplantibacillus plantarum
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2023-04-26 Classification: CELL CYCLE Ligands: SO4, EDO |
 |
Organism: Lactiplantibacillus plantarum
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-04-26 Classification: CELL CYCLE Ligands: TLA |
 |
Organism: Lactiplantibacillus plantarum
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2023-04-26 Classification: CELL CYCLE Ligands: TCE, TLA, GOL |
 |
Organism: Lactiplantibacillus plantarum
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2023-04-26 Classification: CELL CYCLE Ligands: TLA, GOL, G3P |
 |
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2023-02-22 Classification: CELL CYCLE |
 |
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2023-02-22 Classification: CELL CYCLE |
 |
Crystal Structure Of Catalytic Domain Of Lytb (E585Q) From Streptococcus Pneumoniae In Complex With Nag-Nam-Nag-Nam Tetrasaccharide
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-09-21 Classification: HYDROLASE Ligands: 1PE, PEG, CA |
 |
Crystal Structure Of Catalytic Domain In Open Conformation Of Lytb From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: 1PE, PGE, PEG, CA |
 |
Crystal Structure Of Catalytic Domain In Closed Conformation Of Lytb (E585Q)From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: 1PE, PGE, PEG, ACT, CA |
 |
Crystal Structure Of Catalytic Domain Of Lytb From Streptococcus Pneumoniae In Complex With Nag-Nag-Nag-Nag Tetrasaccharide
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: 1PE, PEG, CA |
 |
Crystal Structure Of Catalytic Domain Of Lytb (E585Q) From Streptococcus Pneumoniae In Complex With Nag-Nam-Nag-Nam-Nag Peptidolycan Analogue
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: 1PE, PGE, PEG, CA |
 |
Crystal Structure Of Choline-Binding Module Of Lytb From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: CHT |
 |
Crystal Structure Of Catalytic Domain In Closed Conformation Of Lytb From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae r6
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: 1PE, PEG, CA |
 |
Crystal Structure Of Choline-Binding Module (R1-R9) Of Lytb From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: CHT, ZN, PGE |

