Search Count: 104
 |
Crystal Structure Of Saccharomyces Cerevisiae Nmd4 Protein Bound To Upf1 Helicase Domain
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-08-14 Classification: RNA BINDING PROTEIN Ligands: EDO, SIN, FMT, MLA |
 |
Crystal Structure Of Saccharomyces Cerevisiae Nmd4 Protein Involved In Nonsense Mediated Mrna Decay
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-08-14 Classification: RNA BINDING PROTEIN Ligands: GOL, ACY |
 |
Organism: Leptosphaeria maculans
Method: X-RAY DIFFRACTION Release Date: 2023-03-01 Classification: PROTEIN BINDING |
 |
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2022-03-23 Classification: RNA BINDING PROTEIN Ligands: SO4, GOL |
 |
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2020-12-23 Classification: RNA BINDING PROTEIN Ligands: PEG, EDO, SO4 |
 |
Organism: Schizosaccharomyces pombe 972h-, Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2020-12-23 Classification: RNA BINDING PROTEIN |
 |
Structure Of Archaeoglobus Fulgidus Trm11 M2G10 Trna Methyltransferase Enzyme Bound To Sinefungin
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2020-09-23 Classification: RNA BINDING PROTEIN Ligands: SFG, EDO |
 |
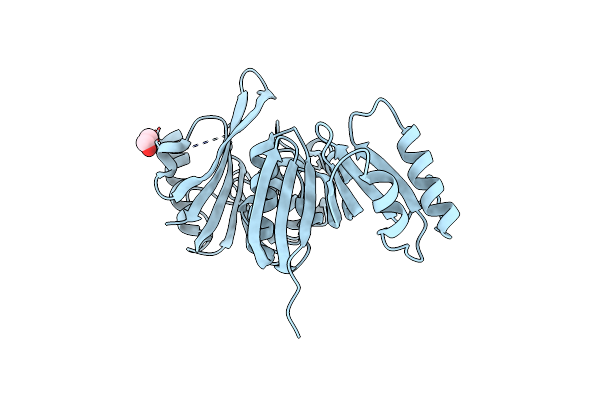
Structure Of Archaeoglobus Fulgidus Trm11-Trm112 M2G10 Trna Methyltransferase Complex Bound To Sinefungin
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2020-09-23 Classification: RNA BINDING PROTEIN Ligands: SFG, EDO, ACY, SO4, ZN |
 |
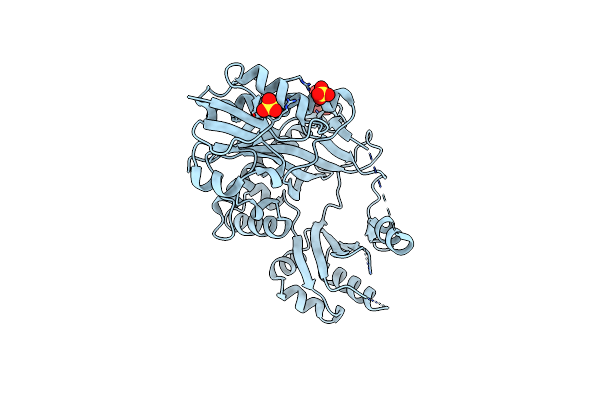
Structure Of Archaeoglobus Fulgidus Trm11 M2G10 Trna Methyltransferase Enzyme
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2020-09-23 Classification: RNA BINDING PROTEIN Ligands: EDO |
 |
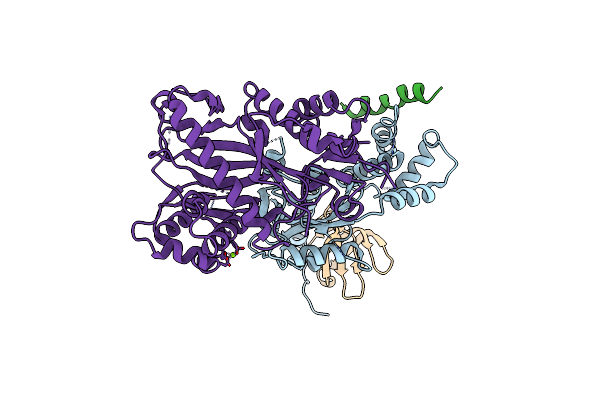
Crystal Structure Of The C-Terminal Domain From K. Lactis Pby1, An Atp-Grasp Enzyme Interacting With The Mrna Decapping Enzyme Dcp2
Organism: Kluyveromyces lactis (strain atcc 8585 / cbs 2359 / dsm 70799 / nbrc 1267 / nrrl y-1140 / wm37)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-04-29 Classification: LIGASE Ligands: SO4 |
 |
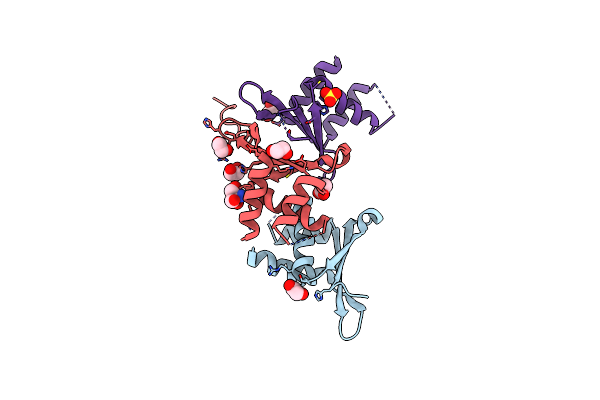
Crystal Structure Of The Pby1 Atp-Grasp Enzyme Bound To The S. Cerevisiae Mrna Decapping Complex (Dcp1-Dcp2-Edc3)
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:3.49 Å Release Date: 2020-04-29 Classification: HYDROLASE Ligands: MG |
 |
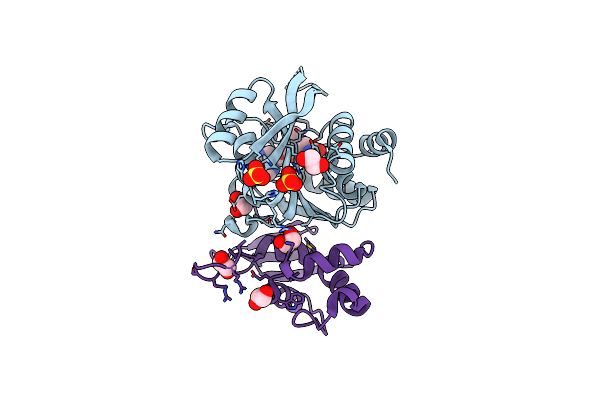
Structure Of S. Pombe Erh1, A Protein Important For Meiotic Mrna Decay In Mitosis And Meiosis Progression.
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-02-12 Classification: CELL CYCLE Ligands: EDO, TRS, ACY, SO4 |
 |
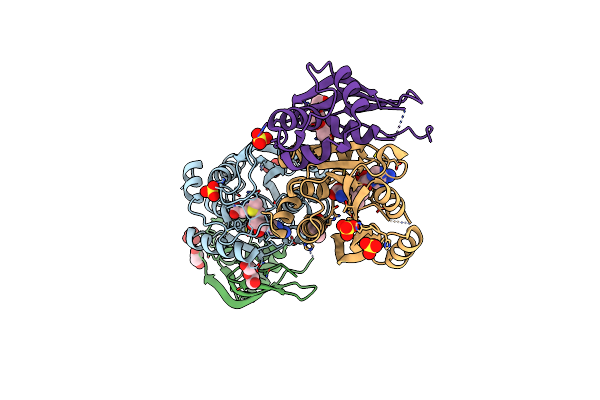
Crystal Structure Of Human Mettl5-Trmt112 Complex, The 18S Rrna M6A1832 Methyltransferase At 1.6A Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-07-31 Classification: TRANSFERASE Ligands: SAM, EDO, SO4 |
 |
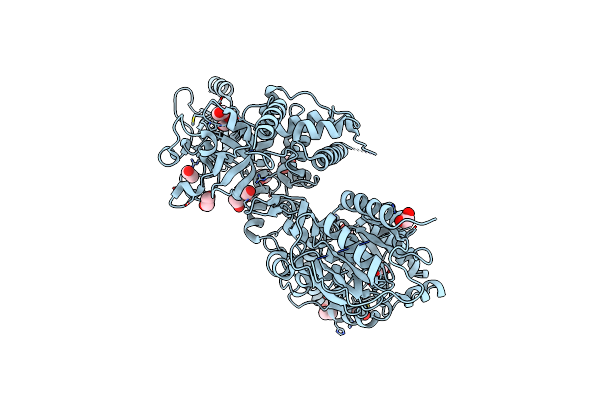
Crystal Structure Of Human Mettl5-Trmt112 Complex, The 18S Rrna M6A1832 Methyltransferase At 2.5A Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2019-07-31 Classification: TRANSFERASE Ligands: SAM, SO4, PEG, EDO, PG0 |
 |
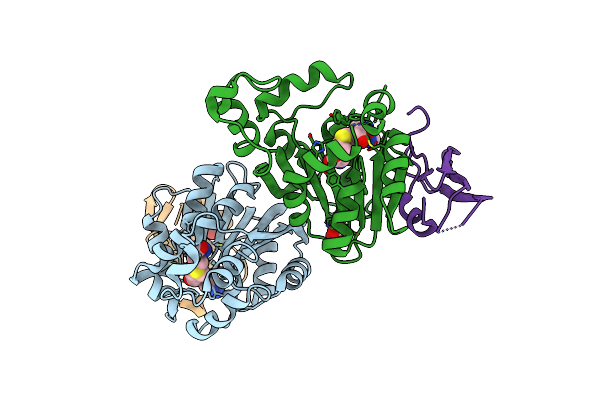
Crystal Structure Of S. Cerevisiae Deah-Box Rna Helicase Dhr1, Essential For Small Ribosomal Subunit Biogenesis
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-06-19 Classification: HYDROLASE Ligands: EDO, MG, CL |
 |
Complex Between The Haloferax Volcanii Trm112 Methyltransferase Activator And The Hvo_0019 Putative Methyltransferase
Organism: Haloferax volcanii, Haloferax volcanii (strain atcc 29605 / dsm 3757 / jcm 8879 / nbrc 14742 / ncimb 2012 / vkm b-1768 / ds2)
Method: X-RAY DIFFRACTION Release Date: 2018-07-11 Classification: TRANSFERASE Ligands: SAH, GOL |
 |
Structure Of C-Terminal Domain From S. Cerevisiae Pat1 Decapping Activator Bound To Dcp2 Hlm2 Peptide (Region 435-451)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-08-16 Classification: RNA BINDING PROTEIN |
 |
Structure Of C-Terminal Domain From S. Cerevisiae Pat1 Decapping Activator Bound To Dcp2 Hlm3 Peptide (Region 484-500)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-08-16 Classification: RNA BINDING PROTEIN Ligands: MG, CL, EDO |
 |
Structure Of C-Terminal Domain From S. Cerevisiae Pat1 Decapping Activator Bound To Dcp2 Hlm10 Peptide (Region 954-970)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2017-08-16 Classification: RNA BINDING PROTEIN Ligands: EDO, MG, TRS |
 |
Organism: Kluyveromyces lactis nrrl y-1140
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2016-10-05 Classification: RNA BINDING PROTEIN |

