Search Count: 23
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-02-13 Classification: METAL BINDING PROTEIN Ligands: GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2017-11-01 Classification: LYASE Ligands: COA, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2017-11-01 Classification: LYASE Ligands: 1VU, EDO, PEG, PGE, PG4, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2017-11-01 Classification: LYASE Ligands: CIT |
 |
Water-Forming Nadh Oxidase From Lactobacillus Brevis (Lbnox) Bound To Nadh.
Organism: Lactobacillus brevis (strain atcc 367 / jcm 1170)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-08-02 Classification: OXIDOREDUCTASE Ligands: FAD, OXY, NAI, EDO |
 |
Crystal Structure Of Engineered Water-Forming Nadph Oxidase (Tpnox) Bound To Nadph. The G159A, D177A, A178R, M179S, P184R Mutant Of Lbnox.
Organism: Lactobacillus brevis kb290
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-08-02 Classification: OXIDOREDUCTASE Ligands: FAD, NDP, EDO, SO4, CL |
 |
Solution Structure Of The Pore-Forming Region Of C. Elegans Mitochondrial Calcium Uniporter (Mcu)
Organism: Caenorhabditis elegans
Method: SOLUTION NMR Release Date: 2016-05-04 Classification: TRANSPORT PROTEIN |
 |
Organism: Lactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2016-04-13 Classification: OXIDOREDUCTASE Ligands: FAD, OXY, EDO |
 |
Structure Of Mn2+-Bound N-Terminal Domain Of Calmodulin In The Presence Of Zn2+
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-08-08 Classification: CALCIUM-BINDING PROTEIN Ligands: MN, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2012-08-08 Classification: CALCIUM-BINDING PROTEIN Ligands: MG |
 |
Structure Of Mg2+ Bound N-Terminal Domain Of Calmodulin In The Presence Of Zn2+
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-08-08 Classification: CALCIUM-BINDING PROTEIN |
 |
Organism: Oryctolagus cuniculus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2005-11-01 Classification: STRUCTURAL PROTEIN Ligands: CA, ATP, GOL, MG, FMT |
 |
Organism: Oryctolagus cuniculus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2005-11-01 Classification: STRUCTURAL PROTEIN Ligands: CA, ATP, GOL, MG |
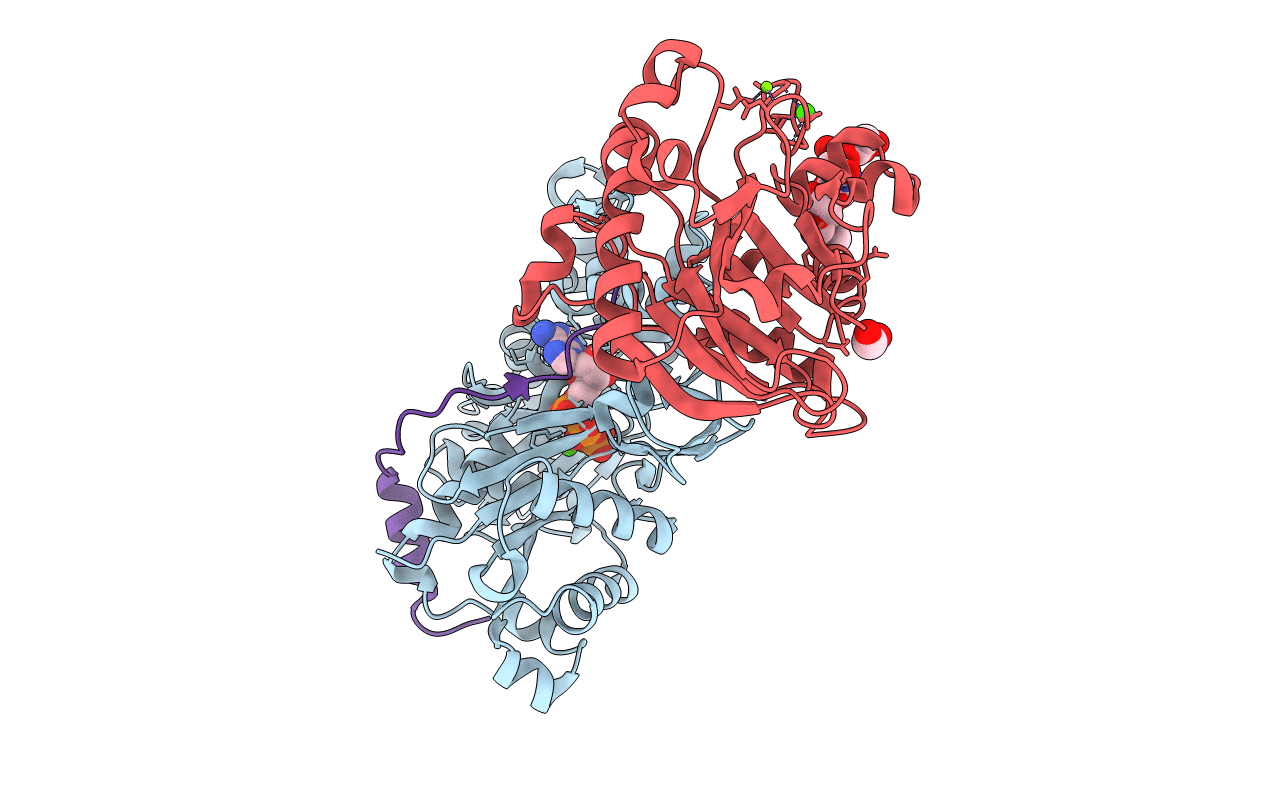 |
Organism: Oryctolagus cuniculus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2005-11-01 Classification: STRUCTURAL PROTEIN Ligands: CA, ATP, MG, FMT |
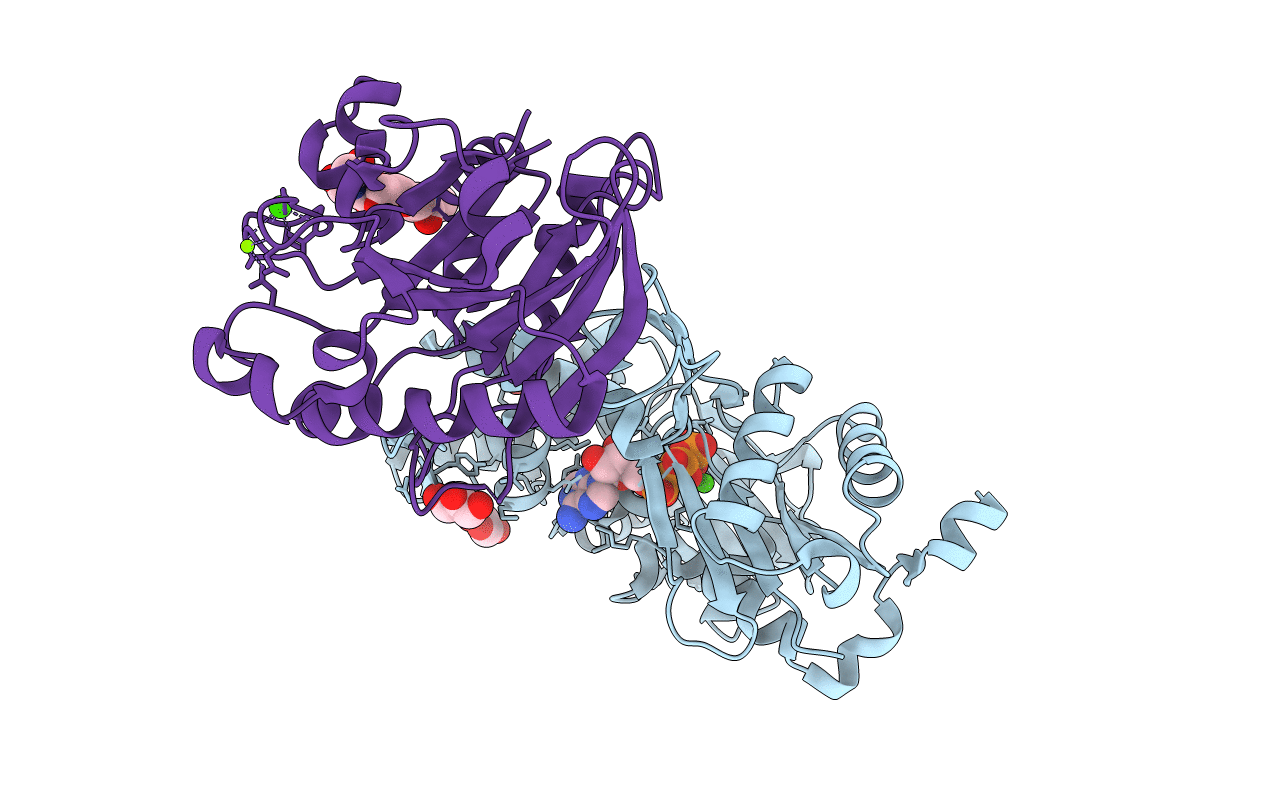 |
Organism: Oryctolagus cuniculus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2005-11-01 Classification: STRUCTURAL PROTEIN Ligands: CA, ATP, GOL, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2005-03-01 Classification: Calcium-Binding Protein Ligands: CA, MPD, TBU |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-11-04 Classification: CELL CYCLE |
 |
Crystal Structure Of The Adenylyl Cyclase Domain Of Anthrax Edema Factor (Ef) In Complex With Calmodulin And 2' Deoxy, 3' Anthraniloyl Atp
Organism: Bacillus anthracis, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2002-12-04 Classification: LYASE Ligands: YB, DOT, CA |
 |
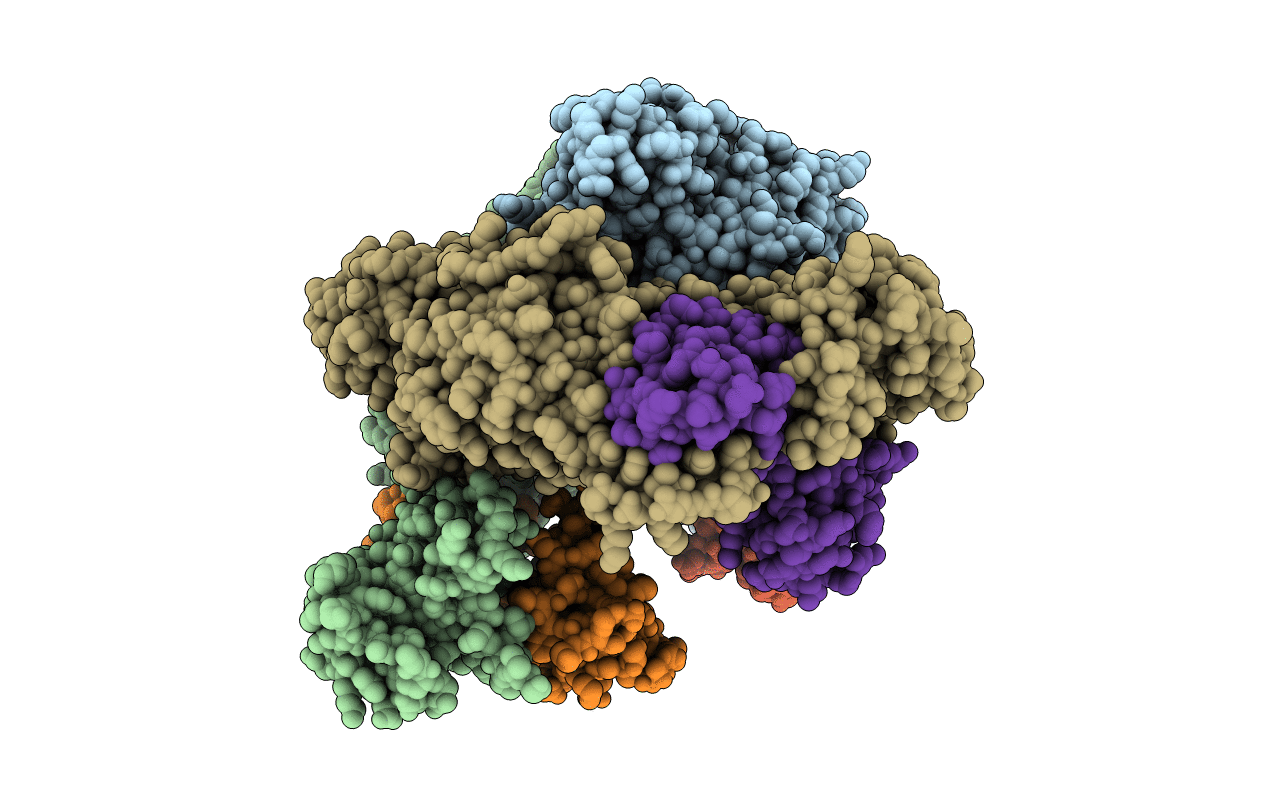
Crystal Structure Of The Adenylyl Cyclase Domain Of Anthrax Edema Factor (Ef)
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2002-01-23 Classification: TOXIN,LYASE Ligands: SO4, NI |
 |
Crystal Structure Of The Adenylyl Cyclase Domain Of Anthrax Edema Factor (Ef) In Complex With Calmodulin And 3' Deoxy-Atp
Organism: Bacillus anthracis, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2002-01-23 Classification: TOXIN,LYASE/METAL BINDING PROTEIN Ligands: YB, 3AT, CA |

