Search Count: 51
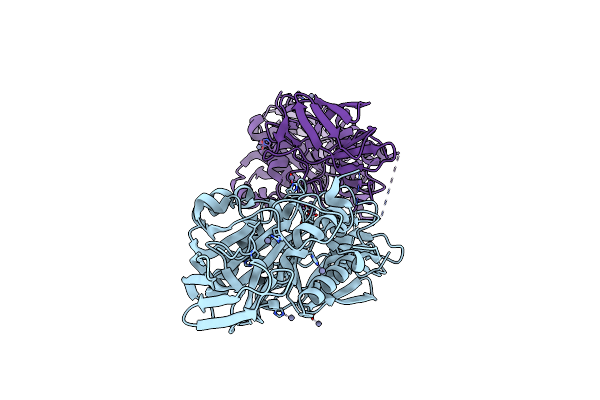 |
Crystal Structure Of Xac1771, A Novel Carbohydrate Acetylesterase From Xanthomonas Citri
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-05-26 Classification: HYDROLASE Ligands: ZN |
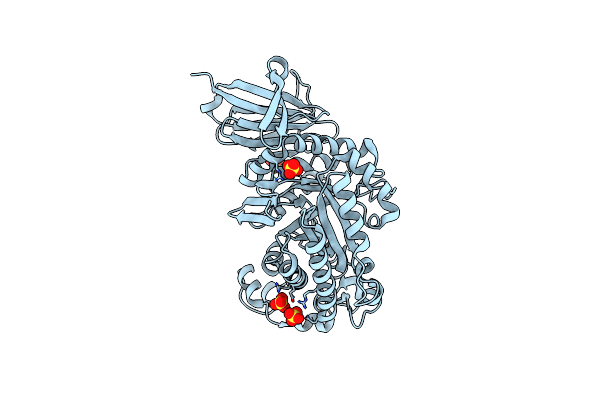 |
Crystal Structure Of Xac1772, A Gh35 Xyloglucan-Active Beta-Galactosidase From Xanthomonas Citri
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-05-26 Classification: HYDROLASE Ligands: SO4 |
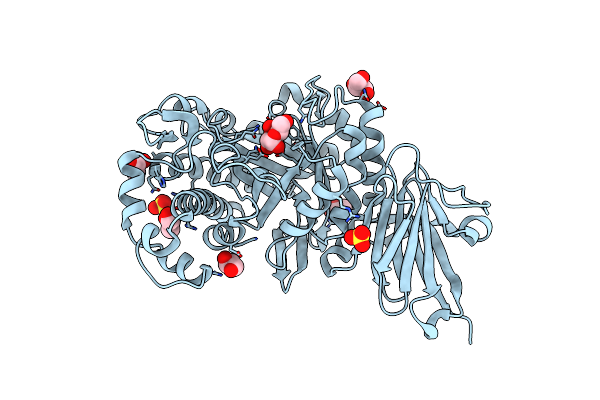 |
Crystal Structure Of The Gh35 Beta-Galactosidase (Xac1772) From Xanthomonas Citri In Complex With Galactose
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-05-26 Classification: HYDROLASE Ligands: SO4, GLA, GOL |
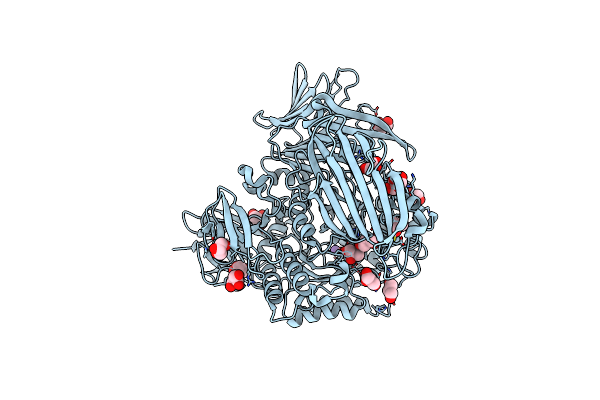 |
Crystal Structure Of The Gh31 Alpha-Xylosidase (Xac1773) From Xanthomonas Citri
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2021-05-26 Classification: HYDROLASE Ligands: GOL, K |
 |
Crystal Structure Of The Gh95 Alpha-L-1,2-Fucosidase (Xac1774) From Xanthomonas Citri
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2021-05-26 Classification: HYDROLASE Ligands: GOL, CA |
 |
Crystal Structure Of The Gh74 Xyloglucanase From Xanthomonas Campestris (Xcc1752)
Organism: Xanthomonas campestris pv. campestris (strain atcc 33913 / dsm 3586 / ncppb 528 / lmg 568 / p 25)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2021-05-26 Classification: HYDROLASE Ligands: EDO, NA, IOD |
 |
Crystal Structure Of The Gh31 Alpha-Xylosidase (Xac1773) From Xanthomonas Citri
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2021-05-26 Classification: HYDROLASE Ligands: K, XYS, GOL |
 |
Organism: Unidentified
Method: POWDER DIFFRACTION Release Date: 2020-12-23 Classification: DE NOVO PROTEIN Ligands: OXL |
 |
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: CA, GOL, PEG |
 |
Crystal Structure Of The Gh43_1 Enzyme From Xanthomonas Citri Complexed With Xylose
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: CA, XYP, PEG, GOL |
 |
Crystal Structure Of The Gh43_1 Enzyme From Xanthomonas Citri Complexed With Xylotriose
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: CA, XYP, PEG, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2020-11-11 Classification: HORMONE Ligands: ZN, PO4, HC4, SCN |
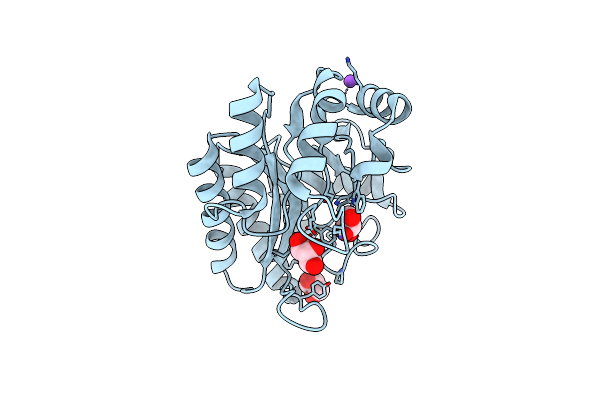 |
Crystal Structure Of A Gh128 (Subgroup I) Endo-Beta-1,3-Glucanase From Amycolatopsis Mediterranei (Amgh128_I)
Organism: Amycolatopsis mediterranei
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: GOL, NA |
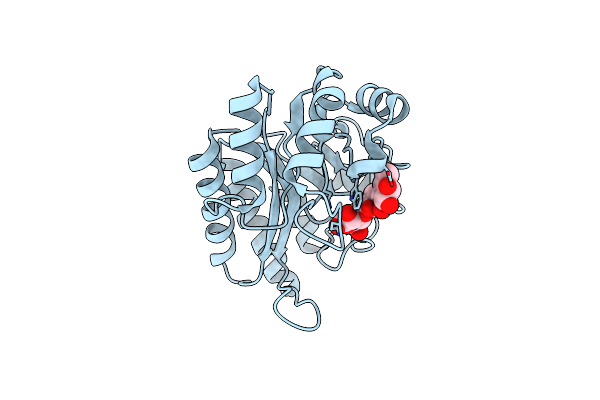 |
Crystal Structure Of A Gh128 (Subgroup I) Endo-Beta-1,3-Glucanase From Amycolatopsis Mediterranei (Amgh128_I) In Complex With Laminaritriose
Organism: Amycolatopsis mediterranei
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-05-20 Classification: HYDROLASE |
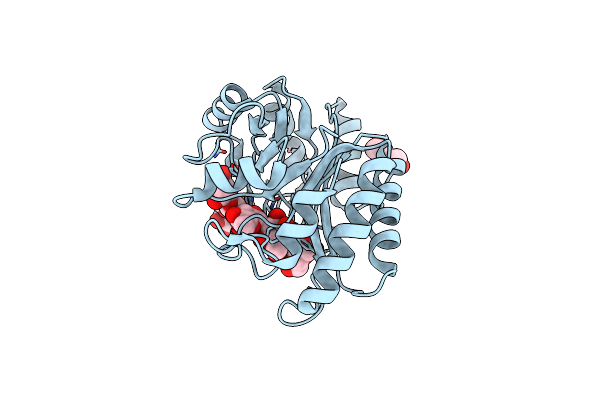 |
Crystal Structure Of A Gh128 (Subgroup I) Endo-Beta-1,3-Glucanase (E199A Mutant) From Amycolatopsis Mediterranei (Amgh128_I) In Complex With Laminaripentaose
Organism: Amycolatopsis mediterranei
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: BGC, ZN, PEG |
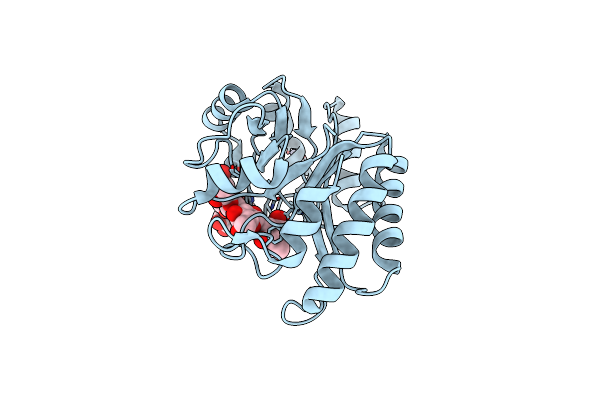 |
Crystal Structure Of A Gh128 (Subgroup I) Endo-Beta-1,3-Glucanase (E102A Mutant) From Amycolatopsis Mediterranei (Amgh128_I) In Complex With Laminaripentaose
Organism: Amycolatopsis mediterranei
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of A Gh128 (Subgroup I) Endo-Beta-1,3-Glucanase (E102A Mutant) From Amycolatopsis Mediterranei (Amgh128_I) In Complex With Laminaritriose And Laminaribiose
Organism: Amycolatopsis mediterranei
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: ZN, PEG |
 |
Crystal Structure Of A Gh128 (Subgroup Ii) Endo-Beta-1,3-Glucanase From Pseudomonas Viridiflava (Pvgh128_Ii)
Organism: Pseudomonas viridiflava
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: SO4, GOL |
 |
Crystal Structure Of A Gh128 (Subgroup Ii) Endo-Beta-1,3-Glucanase From Pseudomonas Viridiflava (Pvgh128_Ii) In Complex With Laminaritriose
Organism: Pseudomonas viridiflava
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: SO4 |
 |
Crystal Structure Of A Gh128 (Subgroup Ii) Endo-Beta-1,3-Glucanase From Sorangium Cellulosum (Scgh128_Ii)
Organism: Sorangium cellulosum so ce56
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: TRS, EPE, CA, CL |

