Search Count: 42
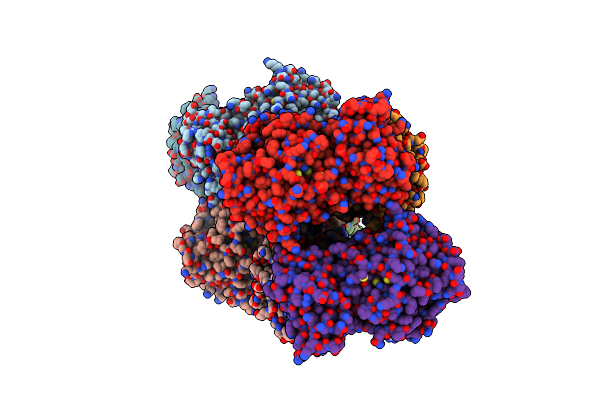 |
Structure Of Gh43 Family Enzyme, Xylan 1, 4 Beta- Xylosidase From Pseudopedobacter Saltans
Organism: Pseudopedobacter saltans dsm 12145
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2023-11-15 Classification: HYDROLASE Ligands: CA |
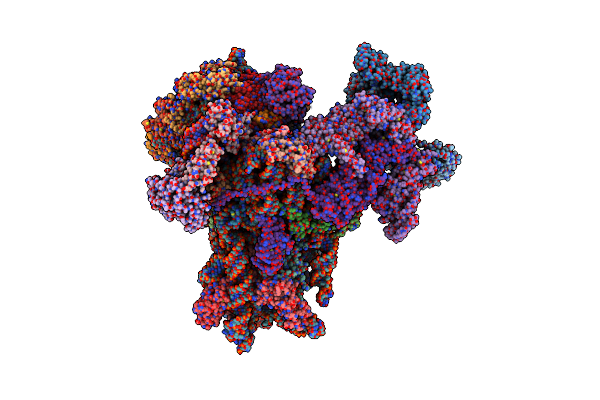 |
Structure Of The Human 48S Initiation Complex In Open State (H48S Aug Open)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-05-11 Classification: RIBOSOME Ligands: ZN, MG |
 |
Structure Of The Human 48S Initiation Complex In Closed State (H48S Aug Closed)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-05-11 Classification: RIBOSOME Ligands: ZN |
 |
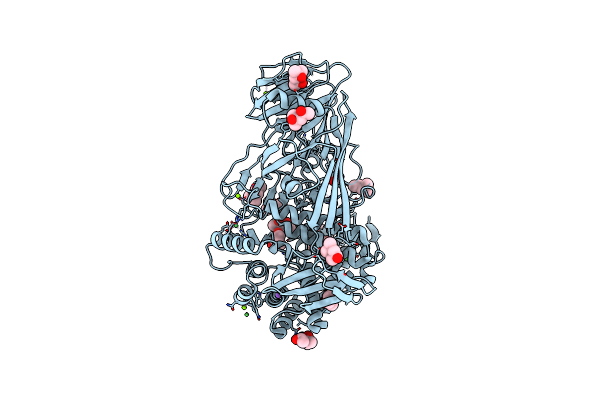
Inhibitor Bound Crystal Structure Of N-Terminal Domain Of Facl13 From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis cdc1551
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2019-04-24 Classification: LIGASE Ligands: JSA |
 |
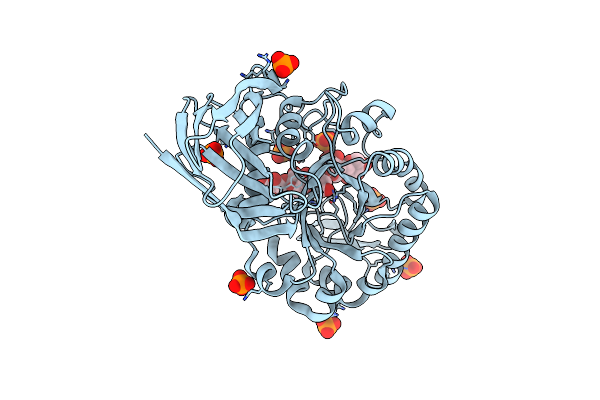
Glycoside Hydrolase Family 81 From Clostridium Thermocellum (Ctlam81A), Mutant E515A
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-03-13 Classification: HYDROLASE Ligands: CA, MPD, CL, NA, ACT, MG, NI |
 |
High Resolution Structure Of The Thermostable Glucuronoxylan Endo-Beta-1, 4-Xylanase, Ctxyn30A, From Clostridium Thermocellum With Two Xylobiose Units Bound
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-10-19 Classification: HYDROLASE Ligands: PO4, PEG |
 |
Determining The Specificities Of The Catalytic Site From The Very High Resolution Structure Of The Thermostable Glucuronoxylan Endo-Beta-1, 4-Xylanase, Ctxyn30A, From Clostridium Thermocellum With A Xylotetraose Bound
Organism: Ruminiclostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2016-10-19 Classification: HYDROLASE Ligands: PO4, PEG |
 |
The Ultra High Resolution Structure Of A Novel Alpha-L-Arabinofuranosidase (Ctgh43) From Clostridium Thermocellum Atcc 27405 With Bound Trimethyl N-Oxide (Trs)
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:0.97 Å Release Date: 2016-07-27 Classification: HYDROLASE Ligands: CA, TRS |
 |
The High Resolution Structure Of A Novel Alpha-L-Arabinofuranosidase (Ctgh43) From Clostridium Thermocellum Atcc 27405
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2016-07-27 Classification: HYDROLASE Ligands: SO4, ACT, GOL |
 |
Structure Of A Novel Carbohydrate Binding Module From Ruminococcus Flavefaciens Fd-1 Endoglucanase Cel5A Solved At The As Edge
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2016-06-29 Classification: SUGAR BINDING PROTEIN Ligands: CAC, GOL |
 |
Very High Resolution Structure Of A Novel Carbohydrate Binding Module From Ruminococcus Flavefaciens Fd-1 Endoglucanase Cel5A
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2016-06-22 Classification: SUGAR BINDING PROTEN Ligands: CAC, GOL |
 |
The Complexity Of The Ruminococcus Flavefaciens Cellulosome Reflects An Expansion In Glycan Recognition
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2016-06-22 Classification: SUGAR BINDING PROTEIN Ligands: CA, NA |
 |
The Complexity Of The Ruminococcus Flavefaciens Cellulosome Reflects An Expansion In Glycan Recognition
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2016-06-22 Classification: SUGAR BINDING PROTEIN |
 |
The Complexity Of The Ruminococcus Flavefaciens Cellulosome Reflects An Expansion In Glycan Recognition
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-06-22 Classification: SUGAR BINDING PROTEIN |
 |
The Complexity Of The Ruminococcus Flavefaciens Cellulosome Reflects An Expansion In Glycan Recognition
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-06-22 Classification: SUGAR BINDING PROTEIN |
 |
Semet Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 9 (Cel9A) From Ruminococcus Flavefaciens Fd-1 In The Orthorhombic Form
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN Ligands: 2PE, P6G, EDO, GOL, PEG, CA, PG4, HHD |
 |
Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 5 Glucanase From Ruminococcus Flavefaciens Fd-1
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN |
 |
Semet Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 5 Glucanase From Ruminococcus Flavefaciens Fd-1
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN |
 |
Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 5 Glucanase From Ruminococcus Flavefaciens Fd-1 Collected At The Zn Edge
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN |
 |
Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 5 Glucanase From Ruminococcus Flavefaciens Fd-1 At Medium Resolution
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN |

