Search Count: 16
 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2021-06-30 Classification: HYDROLASE Ligands: ATP, EDO, GOL |
 |
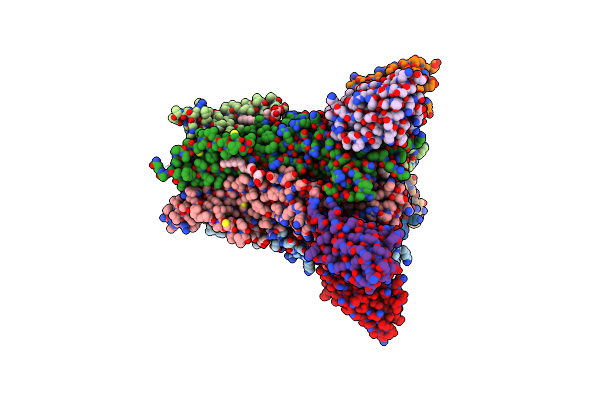
Lmrp From L. Lactis, In An Outward-Open Conformation, Bound To Hoechst 33342
Organism: Lactococcus lactis
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2020-07-29 Classification: TRANSPORT PROTEIN Ligands: HT1, XP4, LMU, GOL |
 |
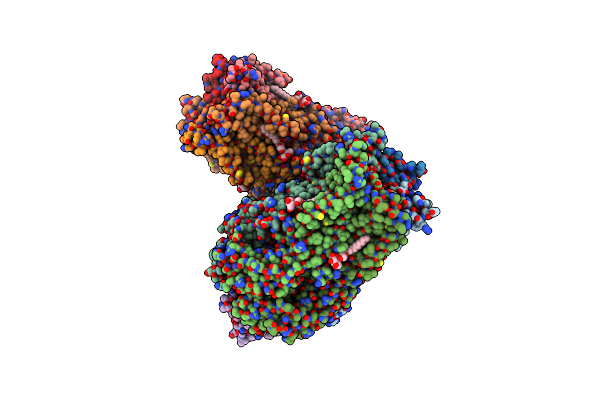
Structure Of The Pentameric Ligand-Gated Ion Channel Elic In Complex With A Pam Nanobody
Organism: Dickeya chrysanthemi, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2020-02-12 Classification: TRANSPORT PROTEIN Ligands: ABU, MES, GOL, CA, PG4, UMQ, ACT |
 |
Structure Of The Pentameric Ligand-Gated Ion Channel Elic In Complex With A Nam Nanobody
Organism: Dickeya chrysanthemi, Lama glama
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2020-02-12 Classification: TRANSPORT PROTEIN Ligands: CA, UMQ |
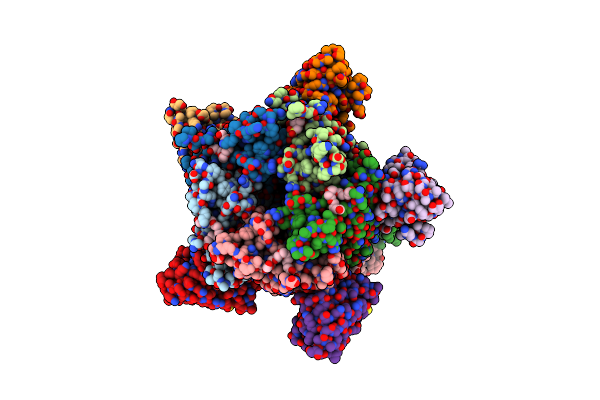 |
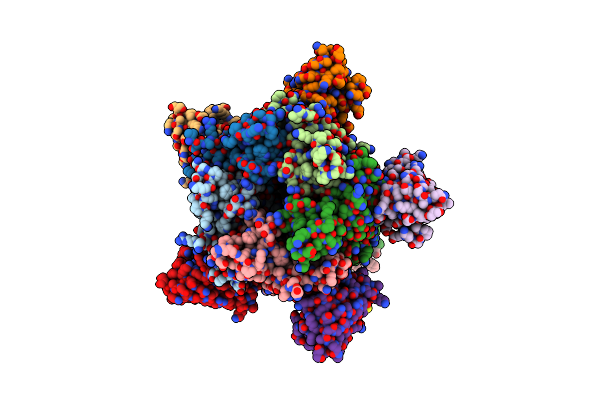
X-Ray Structure Of A Pentameric Ligand Gated Ion Channel From Erwinia Chrysanthemi (Elic) 7'C Pore Mutant (L238C) In Complex With Nanobody 72
Organism: Dickeya chrysanthemi, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-10-09 Classification: MEMBRANE PROTEIN Ligands: P6G, PTY, LMT, MES, NA |
 |
X-Ray Structure Of A Pentameric Ligand Gated Ion Channel From Erwinia Chrysanthemi (Elic) Delta8 Truncation Mutant In Complex With Nanobody 72
Organism: Dickeya chrysanthemi, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2019-10-09 Classification: MEMBRANE PROTEIN |
 |
X-Ray Structure Of A Pentameric Ligand Gated Ion Channel From Erwinia Chrysanthemi (Elic) F16'S Pore Mutant (F247S) With Alternate M4 Conformation.
Organism: Dickeya chrysanthemi
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2019-10-09 Classification: MEMBRANE PROTEIN Ligands: LMT |
 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-06-26 Classification: IMMUNE SYSTEM Ligands: MG, ATP |
 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-06-26 Classification: HYDROLASE Ligands: GOL, PEG |
 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2019-06-19 Classification: HYDROLASE Ligands: ATP |
 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2019-06-19 Classification: HYDROLASE Ligands: GOL, MG, ATP |
 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2019-06-19 Classification: HYDROLASE Ligands: ATP, MG, GOL |
 |
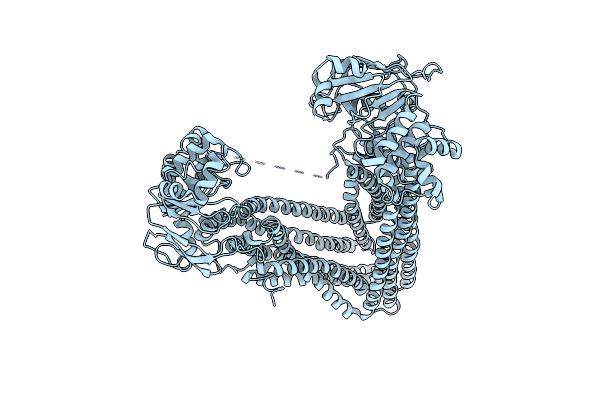
Structures Of P-Glycoprotein Reveal Its Conformational Flexibility And An Epitope On The Nucleotide-Binding Domain
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2013-07-31 Classification: MEMBRANE PROTEIN, TRANSPORT PROTEIN |
 |
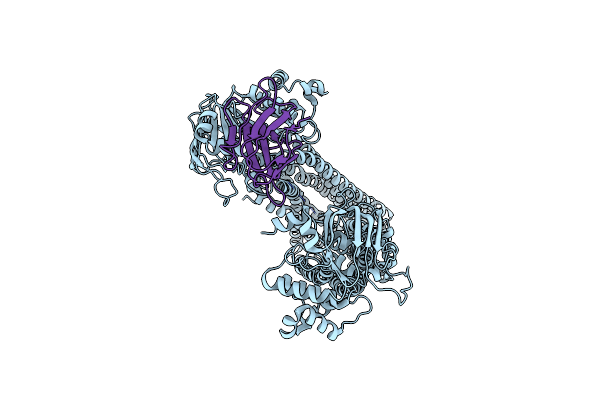
Structures Of P-Glycoprotein Reveal Its Conformational Flexibility And An Epitope On The Nucleotide-Binding Domain
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2013-07-31 Classification: MEMBRANE PROTEIN, TRANSPORT PROTEIN |
 |
Structures Of P-Glycoprotein Reveal Its Conformational Flexibility And An Epitope On The Nucleotide-Binding Domain
Organism: Mus musculus, Lama glama
Method: X-RAY DIFFRACTION Resolution:4.10 Å Release Date: 2013-07-31 Classification: MEMBRANE PROTEIN, TRANSPORT PROTEIN |
 |
Organism: Cupriavidus metallidurans
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2010-06-30 Classification: METAL TRANSPORT Ligands: ZN |

