Search Count: 42
 |
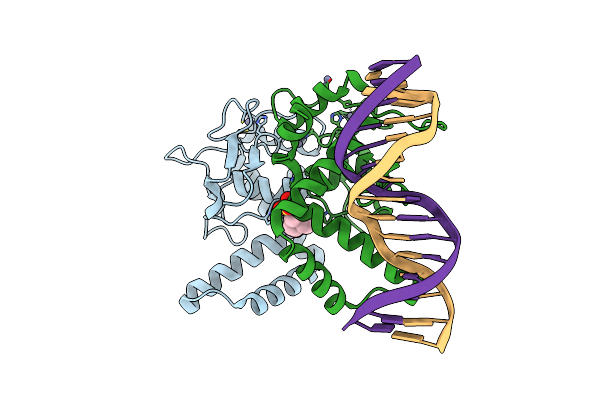
The Structure Of The Dna-Binding Domain Of Nuclear Factor 1 X Bound To Nfi Consensus Dna Sequence
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: TRANSCRIPTION Ligands: ZN |
 |
Structural Determinants Of Cold-Activity And Glucose Tolerance Of A Family 1 Glycoside Hydrolase (Gh1) From Antarctic Marinomonas Ef 1
Organism: Marinomonas sp. ef1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-03-06 Classification: HYDROLASE Ligands: EDO, TRS |
 |
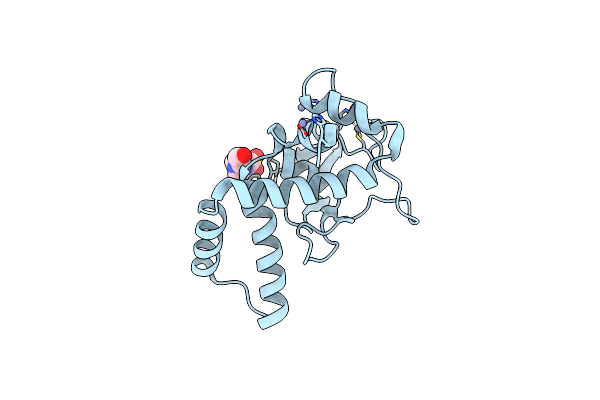
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-01-18 Classification: DNA BINDING PROTEIN Ligands: ZN, EPE |
 |
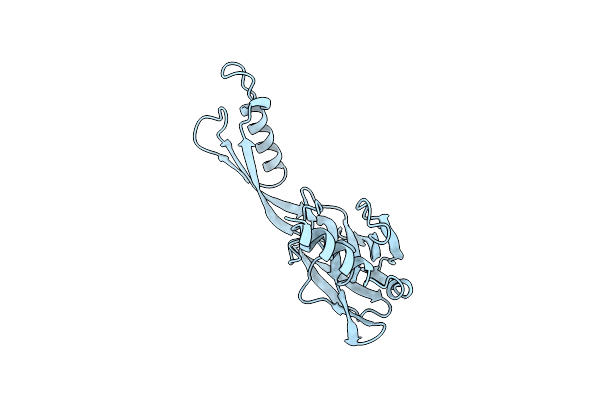
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2023-01-18 Classification: DNA BINDING PROTEIN Ligands: ZN, EPE |
 |
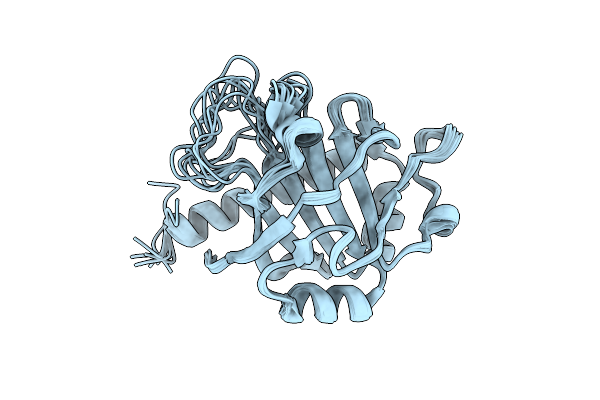
Organism: Trypanosoma cruzi
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2022-02-02 Classification: IMMUNE SYSTEM |
 |
Nmr Solution Structure Of The Sylf Domain Of Burkholderia Pseudomallei Bpsl1445
Organism: Burkholderia pseudomallei
Method: SOLUTION NMR Release Date: 2021-12-29 Classification: LIPID BINDING PROTEIN |
 |
Organism: Serratia sp.
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2021-01-27 Classification: HYDROLASE Ligands: OXD, GOL |
 |
Organism: Schistosoma mansoni
Method: X-RAY DIFFRACTION Resolution:3.22 Å Release Date: 2020-09-30 Classification: ALLERGEN |
 |
Organism: Alicyclobacillus tengchongensis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-07-29 Classification: HYDROLASE Ligands: B3P, EDO, NA, IOD |
 |
The Low Resolution Structure Of A Zinc-Dependent Alcohol Dehydrogenase From Halomonas Elongata.
Organism: Halomonas elongata (strain atcc 33173 / dsm 2581 / nbrc 15536 / ncimb 2198 / 1h9)
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2020-06-17 Classification: OXIDOREDUCTASE Ligands: ZN |
 |
Crystal Structure Of An R-Selective Transaminase From Thermomyces Stellatus.
Organism: Thermomyces stellatus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-04-15 Classification: TRANSFERASE Ligands: EDO, PLP, NA |
 |
Organism: Burkholderia pseudomallei (strain k96243)
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2020-02-19 Classification: HYDROLASE Ligands: MG, CL |
 |
Organism: Burkholderia pseudomallei (strain k96243)
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2020-01-15 Classification: HYDROLASE Ligands: MG, CL |
 |
Organism: Halomonas elongata dsm 2581
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-05-08 Classification: TRANSFERASE Ligands: PLP, EDO, CL |
 |
The Crystal Structure Of A New Transaminase From The Marine Bacterium Virgibacillus
Organism: Virgibacillus pantothenticus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-12-12 Classification: TRANSFERASE Ligands: PLP, EDO, CL |
 |
Organism: Burkholderia pseudomallei k96243
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-12-05 Classification: IMMUNE SYSTEM Ligands: CA, ACT, GOL |
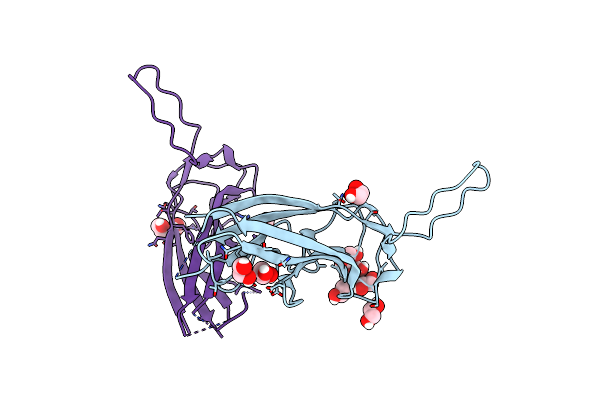 |
The Crystal Structure Of Burkholderia Pseudomallei Antigen And Type I Fimbria Protein Bpsl1626.
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-01-17 Classification: IMMUNE SYSTEM Ligands: EDO |
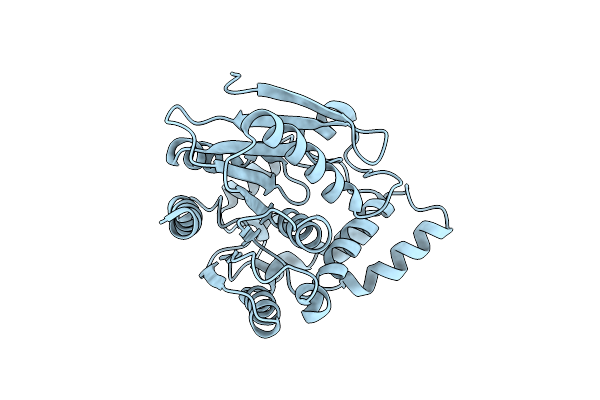 |
The Crystal Structure Of A Highly Thermostable Carboxyl Esterase From Bacillus Coagulans
Organism: Bacillus coagulans dsm 1 = atcc 7050
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-12-06 Classification: HYDROLASE |
 |
The Crystal Structure Of A Highly Thermostable Carboxyl Esterase From Bacillus Coagulans In Complex With Glycerol
Organism: Bacillus coagulans dsm 1 = atcc 7050
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-12-06 Classification: HYDROLASE Ligands: GOL, CL, PEG, ACT |
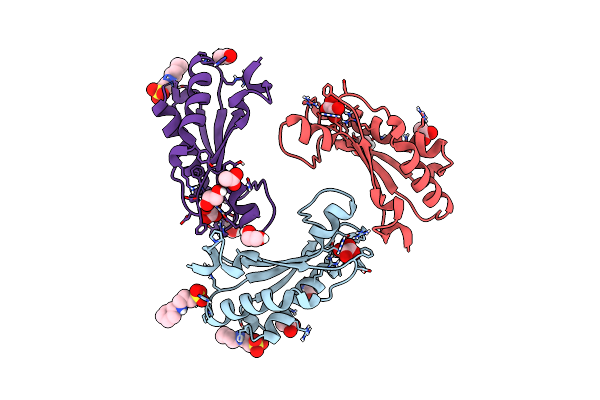 |
Crystal Structure Of The Peptidoglycan-Associated Lipoprotein From Burkholderia Cenocepacia
Organism: Burkholderia cenocepacia j2315
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-07-26 Classification: TRANSPORT PROTEIN Ligands: EDO, CXS, OXL, ACT |

