Search Count: 15
 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro)In Complex With Inhibitor Avi-3318
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: A1BTM |
 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) In Complex With Inhibitor Avi-4692
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE Ligands: A1BVT, A1BTO |
 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro)In Complex With Inhibitor Avi-4516
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: A1BTP, EDO |
 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) Variant Q192T In Complex With Inhibitor Avi-4303
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: EDO, A1BTN, CL |
 |
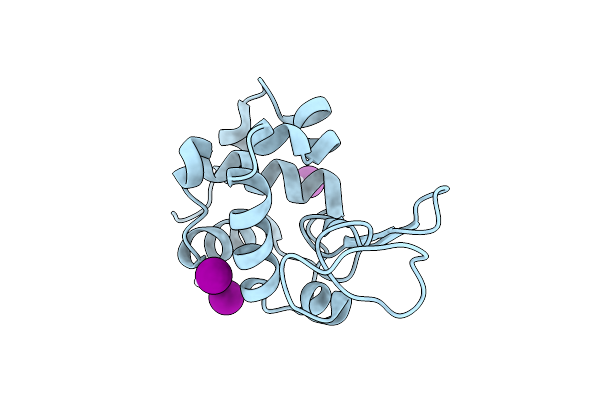
X-Ray Structure Of The Adduct Formed Upon Reaction Of The Diiodido Analogue Of Picoplatin With Lysozyme (Structure A)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2025-05-14 Classification: HYDROLASE Ligands: NH3, ACT, CL, PT, IOD |
 |
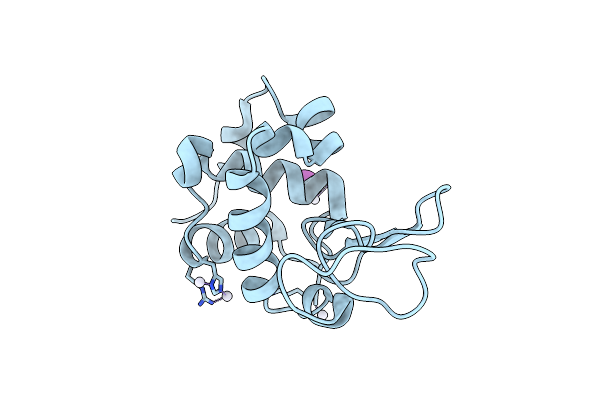
X-Ray Structure Of The Adduct Formed Upon Reaction Of The Diiodido Analogue Of Picoplatin With Lysozyme (Structure B)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2025-05-14 Classification: HYDROLASE Ligands: PT, IOD |
 |
X-Structure Of The Adduct Formed Upon Reaction Of The Diiodido Analogue Of Picoplatin With Lysozyme (Structure C)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2025-05-14 Classification: HYDROLASE Ligands: PT, NH3, IOD |
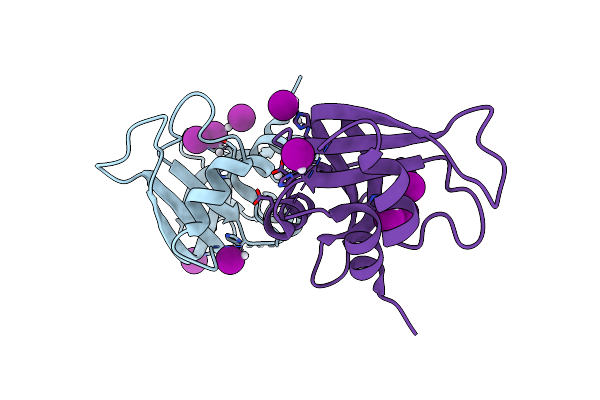 |
X-Ray Structure Of The Adduct Formed Upon Reaction Of The Diiodido Analogue Of Picoplatin With Ribonuclease A
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2025-05-14 Classification: HYDROLASE Ligands: NH3, PT, IOD, CL |
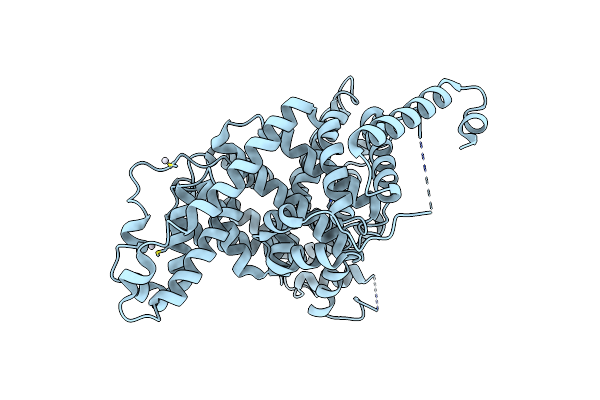 |
X-Ray Structure Of The Adduct Formed Upon Reaction Of The Diiodido Analogue Of Picoplatin With Human Serum Albumin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.90 Å Release Date: 2025-05-14 Classification: HYDROLASE Ligands: PT |
 |
Organism: Lithobates pipiens
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2021-10-06 Classification: HYDROLASE |
 |
Organism: Lithobates pipiens
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2021-10-06 Classification: HYDROLASE Ligands: ACT, SO4 |
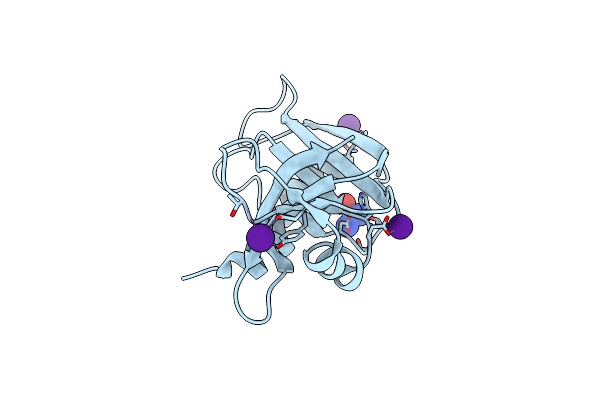 |
The X-Ray Structure Of Bovine Pancreatic Ribonuclease Incubated In The Presence Of An Excess Of Carboplatin (1:10 Ratio)
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2017-08-16 Classification: HYDROLASE Ligands: QPT, CS, CL |
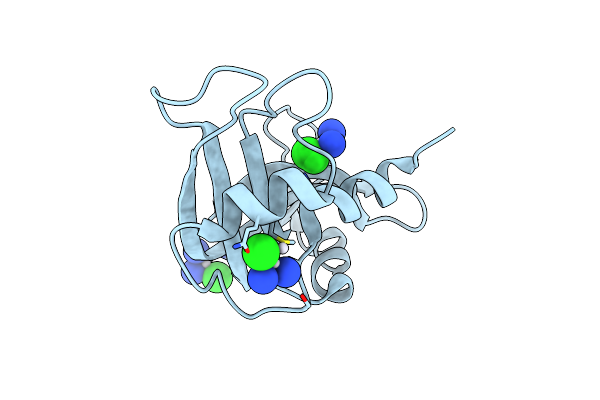 |
The X-Ray Structure Of Bovine Pancreatic Ribonuclease Incubated In The Presence Of An Excess Of Cisplatin (1:10 Ratio)
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2015-03-25 Classification: HYDROLASE Ligands: CPT, CL |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2002-03-13 Classification: HYDROLASE Ligands: SO4 |
 |
Crystal Structure Of An Rnase A Dimer Displaying A New Type Of 3D Domain Swapping
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2001-02-21 Classification: hydrolase/DNA Ligands: PO4, GOL |

