Search Count: 40
 |
Crystal Structure Of Ha3 From Clostridium Botulinum Type B With Alpha2,3-Sialyllactose
Organism: Clostridium botulinum b1 str. okra
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TOXIN |
 |
Crystal Structure Of Ha3 From Clostridium Botulinum Type B With Alpha2,6-Sialyllactose
Organism: Clostridium botulinum b1 str. okra
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TOXIN Ligands: SIA |
 |
Crystal Structure Of Hla-C*12:02 In Complex With Kaynvtqaf (Kf9), A 9-Mer Epitope From Sars-Cov-2 Nucleocapsid (N266-274)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of Gv37-Tcr In Complex With Hla-C*12:02 With Kaynvtqaf (Kf9), A 9-Mer Epitope From Sars-Cov-2 Nucleocapsid (N266-274)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: IMMUNE SYSTEM Ligands: EDO, SO4 |
 |
Cryo-Em Structure Of The Inhibitor-Bound Vo Complex From Enterococcus Hirae
Organism: Enterococcus hirae atcc 9790
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CDL, NA, W3K |
 |
Organism: Microcystis aeruginosa nies-88
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2024-06-19 Classification: TRANSFERASE Ligands: PIS, MG, BTB |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-03-27 Classification: SIGNALING PROTEIN Ligands: ADN |
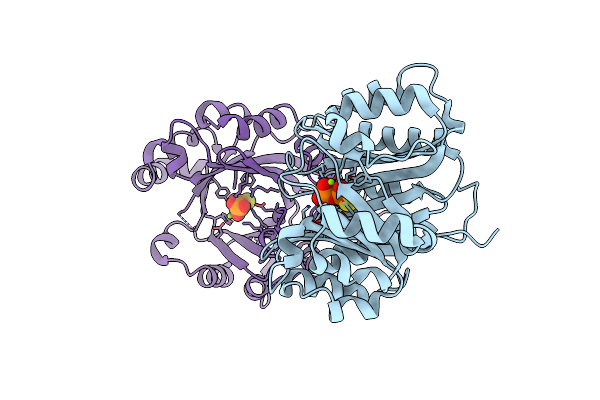 |
Crystal Structure Of Limf Prenyltransferase (H239G/W273T Mutant) Bound With The Thiodiphosphate Moiety Of Farnesyl S-Thiolodiphosphate (Fspp)
Organism: Limnothrix sp. caciam 69d
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2023-11-08 Classification: TRANSFERASE Ligands: MG, PIS |
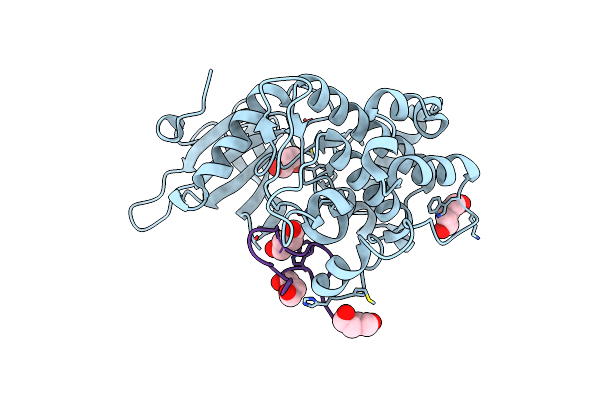 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2022-10-26 Classification: SIGNALING PROTEIN/INHIBITOR Ligands: BU1 |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2022-10-26 Classification: SIGNALING PROTEIN/INHIBITOR Ligands: ANP, MG, SO4, PGE |
 |
Crystal Structure Of Limf Prenyltransferase Bound With A Peptide Substrate And Gspp
Organism: Limnothrix sp. caciam 69d, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2022-08-03 Classification: TRANSFERASE Ligands: MG, GST, PPV |
 |
Organism: Limnothrix sp. caciam 69d
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2022-08-03 Classification: TRANSFERASE Ligands: MG, GST, BTB |
 |
A New V27M Variant Of Beta 2 Microglobulin Induced Amyloidosis In A Patient With Long-Term Hemodialysis
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2021-01-06 Classification: IMMUNE SYSTEM Ligands: GOL, MG, CA, PO4 |
 |
Structure Of 6-Aminohexanoate-Oligomer Hydrolase From Arthrobacter Sp. Ki72.
Organism: Arthrobacter sp.
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-11-21 Classification: HYDROLASE Ligands: GOL, SO4, CL |
 |
Structure Of 6-Aminohexanoate-Oligomer Hydrolase From Arthrobacter Sp. Ki72., D122G Mutant
Organism: Flavobacterium sp. ki723t1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-11-21 Classification: HYDROLASE Ligands: SO4, CL, GOL |
 |
Structure Of 6-Aminohexanoate-Oligomer Hydrolase From Arthrobacter Sp. Ki72., D122R Mutant
Organism: Flavobacterium sp. ki723t1
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2018-11-21 Classification: HYDROLASE Ligands: PO4, GOL |
 |
Structure Of 6-Aminohexanoate-Oligomer Hydrolase From Arthrobacter Sp. Ki72., D122K Mutant
Organism: Flavobacterium sp. ki723t1
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2018-11-21 Classification: HYDROLASE Ligands: PO4, GOL |
 |
Structure Of 6-Aminohexanoate-Oligomer Hydrolase From Arthrobacter Sp. Ki72., D122V Mutant
Organism: Flavobacterium sp. ki723t1
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2018-11-21 Classification: HYDROLASE Ligands: GOL, PO4 |
 |
Structure Of 6-Aminohexanoate-Oligomer Hydrolase From Arthrobacter Sp. Ki72., H130Y Mutant
Organism: Flavobacterium sp. ki723t1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-11-21 Classification: HYDROLASE Ligands: SO4, GOL |
 |
Structure Of 6-Aminohexanoate-Oligomer Hydrolase From Arthrobacter Sp. Ki72., D122G/H130Y Mutant
Organism: Flavobacterium sp. ki723t1
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2018-07-25 Classification: HYDROLASE Ligands: GOL, NA, SO4 |

