Search Count: 24
 |
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Hex In The Ligand-Free Form At 303 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2022-11-09 Classification: TRANSFERASE Ligands: PLP, SO4 |
 |
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Aifs In The Ligand-Free Form At 100 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2022-11-02 Classification: TRANSFERASE Ligands: PLP, SO4 |
 |
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Aifs Bound To Maleic Acid At 100 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2022-11-02 Classification: TRANSFERASE Ligands: MAE, PLP |
 |
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Hex In The Ligand-Free Form At 278 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2022-11-02 Classification: TRANSFERASE Ligands: PLP, SO4 |
 |
Crystal Structure Of Wild-Type E. Coli Aspartate Aminotransferase In The Ligand-Free Form At 278 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2022-11-02 Classification: TRANSFERASE Ligands: PLP, SO4 |
 |
Crystal Structure Of Wild-Type E. Coli Aspartate Aminotransferase Bound To Maleate At 278 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2022-10-05 Classification: TRANSFERASE Ligands: PLP, MAE |
 |
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Vfit In The Ligand-Free Form At 278 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2022-10-05 Classification: TRANSFERASE Ligands: PLP, SO4 |
 |
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Vfit Bound To Maleic Acid At 278 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2022-10-05 Classification: TRANSFERASE Ligands: PLP, MAE |
 |
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Vfiy In The Ligand-Free Form At 278 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2022-10-05 Classification: TRANSFERASE Ligands: SO4, PLP |
 |
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Vfiy Bound To Maleic Acid At 278 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2022-10-05 Classification: TRANSFERASE Ligands: PLP, MAE |
 |
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Hex Bound To Maleic Acid At 278 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-10-05 Classification: TRANSFERASE Ligands: PLP, MAE |
 |
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Vfcs In The Ligand-Free Form At 278 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-10-05 Classification: TRANSFERASE Ligands: SO4, PLP |
 |
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Vfcs Bound To Maleic Acid At 278 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-10-05 Classification: TRANSFERASE Ligands: PLP, MAE |
 |
Crystal Structure Of Wild-Type E. Coli Aspartate Aminotransferase In The Ligand-Free Form At 303 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2022-10-05 Classification: TRANSFERASE Ligands: PLP, SO4 |
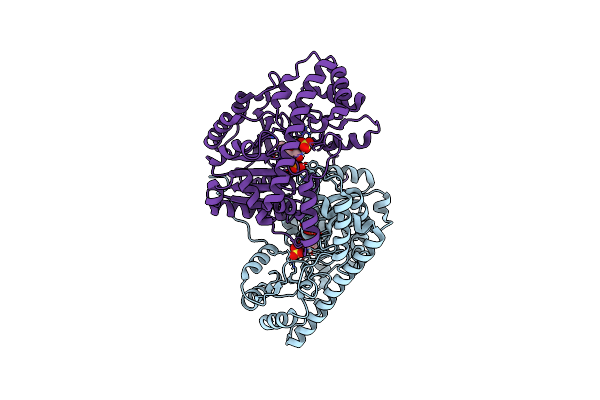 |
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Vfit In The Ligand-Free Form At 303 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2022-10-05 Classification: TRANSFERASE Ligands: PLP, SO4 |
 |
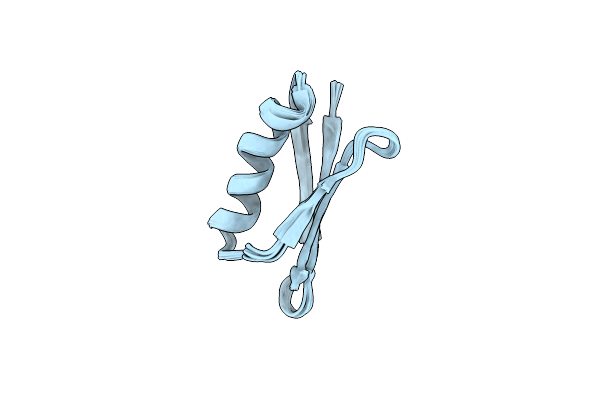
Solution Nmr Structure Of Dancer3-F34A, A Rigid And Natively Folded Single Mutant Of The Dynamic Protein Dancer-3
Organism: Streptococcus sp. group g
Method: SOLUTION NMR Release Date: 2019-08-21 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of Nerd-C, A Natively Folded Tetramutant Of The B1 Domain Of Streptococcal Protein G (Gb1)
Organism: Streptococcus sp. gx7805
Method: SOLUTION NMR Release Date: 2017-08-23 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of Nerd-S, A Natively Folded Pentamutant Of The B1 Domain Of Streptococcal Protein G (Gb1) With A Solvent-Exposed Trp43
Organism: Streptococcus sp. gx7805
Method: SOLUTION NMR Release Date: 2017-08-23 Classification: DE NOVO PROTEIN |
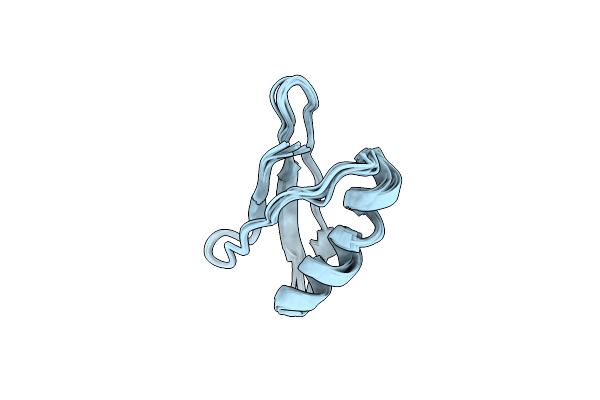 |
Solution Nmr Structure Of The Major Species Of Dancer-2, A Dynamic And Natively Folded Pentamutant Of The B1 Domain Of Streptococcal Protein G (Gb1)
Organism: Streptococcus sp. gx7805
Method: SOLUTION NMR Release Date: 2017-08-23 Classification: DE NOVO PROTEIN |
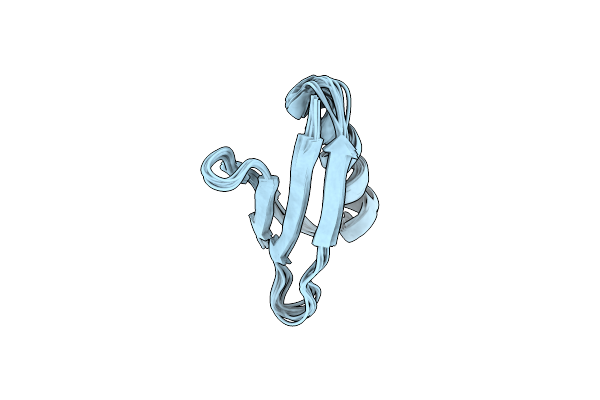 |
Solution Nmr-Derived Model Of The Minor Species Of Dancer-2, A Dynamic And Natively Folded Pentamutant Of The B1 Domain Of Streptococcal Protein G (Gb1)
Organism: Streptococcus sp. gx7805
Method: SOLUTION NMR Release Date: 2017-08-23 Classification: DE NOVO PROTEIN |

