Search Count: 23
 |
Organism: Aspergillus niger
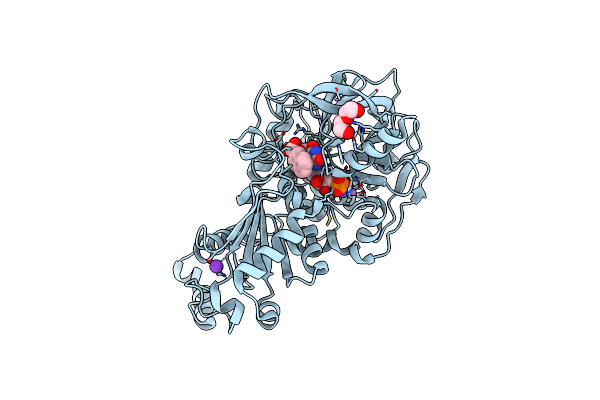
Method: X-RAY DIFFRACTION Resolution:1.07 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, BYN, FIV, PEG |
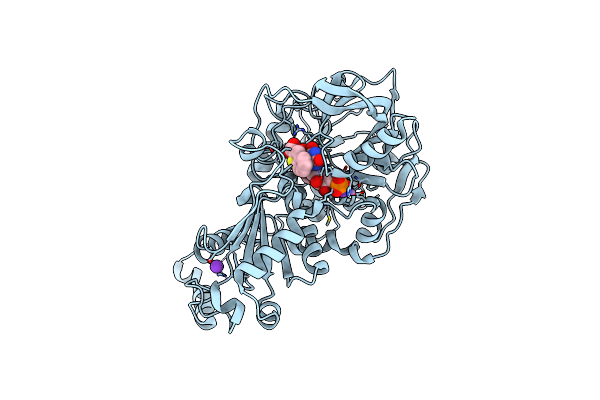 |
Structure Of A. Niger Fdc I327S Variant In Complex With Benzothiophene 2 Carboxylic Acid
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, BYN, ND8 |
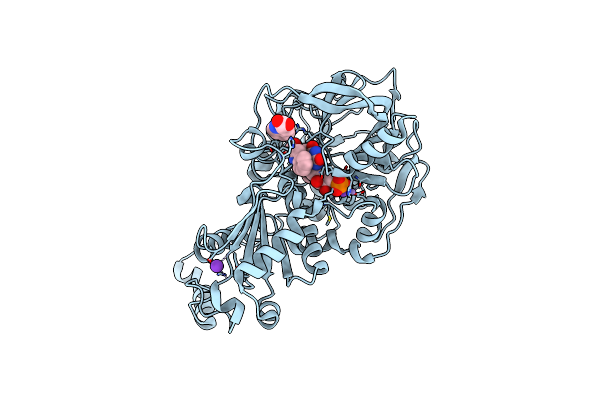 |
Structure Of A. Niger Fdc I327S Variant In Complex With Indol-2-Carboxylic Acid
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, BYN, ICB |
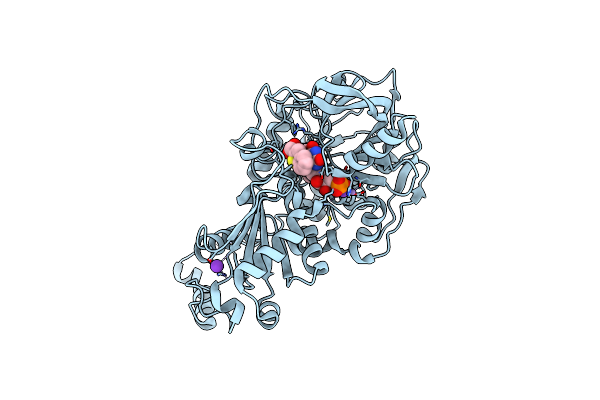 |
Structure Of A. Niger Fdc Wt In Complex With Benzothiophene 2 Carboxylic Acid
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, BYN, ND8 |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, BYN, ICB |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, FMN, FIV |
 |
Structure Of A. Niger Fdc Wt In Complex With Fmn And Indole 2 Carboxylic Acid
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, FMN, ICB |
 |
Structure Of A. Niger Fdc Wt In Complex With Fmn And Benzothiophene 2 Carboxylic Acid
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, FMN, ND8 |
 |
Aspergillus Niger Ferulic Acid Decarboxylase (Fdc) In Complex With Fmn And Cinnamic Acid
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2019-08-28 Classification: LYASE Ligands: FMN, MN, K, TCA |
 |
Aspergillus Niger Ferulic Acid Decarboxylase (Fdc) In Complex With Prfmn (Purified In Dark) And Alphafluorocinnamic Acid
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Release Date: 2019-08-28 Classification: LYASE Ligands: MN, K, BYN, 4LW |
 |
Aspergillus Niger Ferulic Acid Decarboxylase (Fdc) In Complex With Prfmn (Purified In The Radical Form) And Phenylpropiolic Acid
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2019-08-28 Classification: LYASE Ligands: 4LU, MN, K, JQ8 |
 |
Aspergillus Niger Ferulic Acid Decarboxylase (Fdc) F437L Variant In Complex With The Covalent Adduct Formed Between Prfmn Cofactor And Phenylpropiolic Acid (Int1')
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2019-08-28 Classification: LYASE Ligands: MN, K, JQZ |
 |
Aspergillus Niger Ferulic Acid Decarboxylase (Fdc) L439G Variant In Complex With The Covalent Adduct Formed Between Prfmn Cofactor And Phenylpropiolic Acid (Int1')
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2019-08-28 Classification: LYASE Ligands: JQZ, MN, K, JQ8 |
 |
Aspergillus Niger Ferrulic Acid Decarboxylase (Fdc) In Complex With The Covalent Adduct Formed Between Prfmn Cofactor And Phenylpropiolic Acid (Int1')
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2019-08-28 Classification: LYASE Ligands: MN, K, JQZ |
 |
Aspergillus Niger Ferulic Acid Decarboxylase (Fdc)In Complex With The Covalent Adduct Formed Between Prfmn Cofactor And Phenylpropiolic Acid, Following Decarboxylation (Int3')
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.01 Å Release Date: 2019-08-28 Classification: LYASE Ligands: MN, K, JQH |
 |
Aspergillus Niger Ferulic Acid Decarboxylase (Fdc) In Complex With The Covalent Adduct Formed Between Prfmn Cofactor And Phenyl Acetylene (Int3')
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2019-08-28 Classification: LYASE Ligands: MN, K, JQH |
 |
Aspergillus Niger Ferulic Acid Decarboxylase (Fdc) E282Q Variant In Complex With The Covalent Adduct Formed Between Prfmn Cofactor And Cinnamic Acid (Int2)
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2019-08-28 Classification: LYASE Ligands: MN, K, JQK |
 |
Aspergillus Niger Ferulic Acid Decarboxylase (Fdc) E282Q Variant In Complex With The Covalent Adduct Formed Between Prfmn Cofactor And Alphafluoro-Cinnamic Acid (Int2)
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2019-08-28 Classification: LYASE Ligands: MN, K, JQQ |
 |
Aspergillus Niger Ferulic Acid Decarboxylase (Fdc) E282Q Variant In Complex With The Covalent Adduct Formed Between Prfmn Cofactor And Pentafluorocinnamic Acid (Int2)
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2019-08-28 Classification: LYASE Ligands: MN, K, JRN |
 |
Aspergillus Niger Ferulic Acid Decarboxylase (Fdc) In Complex With The Covalent Adduct Formed Between Prfmn Cofactor And Crotonic Acid Following Decarboxylation (Int3)
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2019-08-28 Classification: LYASE Ligands: JRH, MN, K |

