Search Count: 13
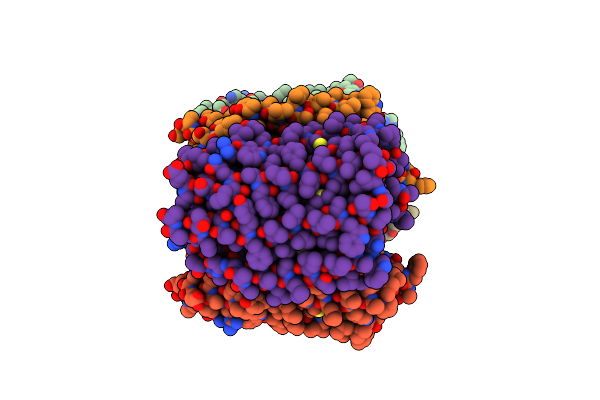 |
Crystal Structure Of The Microbial Rhodopsin From Sphingomonas Paucimobilis (Spar)
Organism: Sphingomonas paucimobilis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-05-03 Classification: MEMBRANE PROTEIN Ligands: LFA |
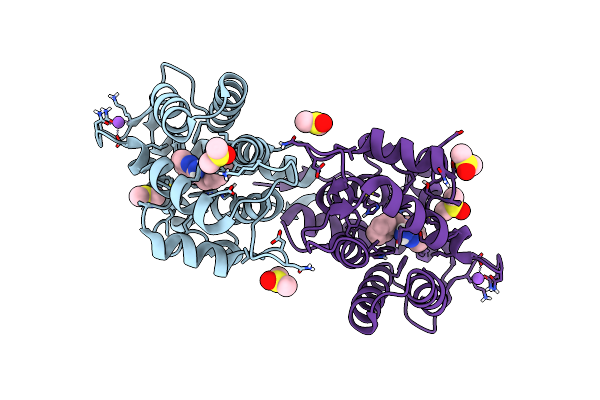 |
Crystal Structure Of Ca2+-Discharged Obelin In Complex With Coelenteramine-V
Organism: Obelia longissima
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-10-19 Classification: LUMINESCENT PROTEIN Ligands: LK9, NA, DMS |
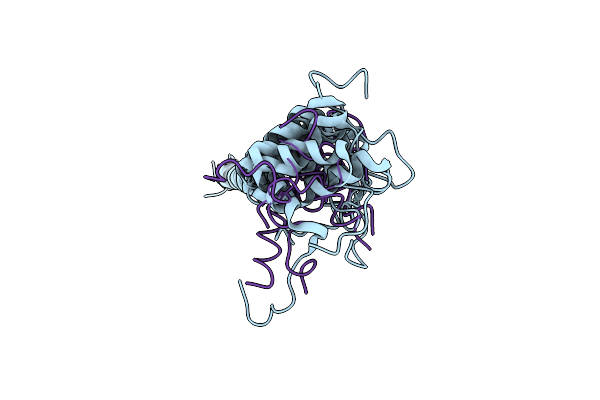 |
Dynamic Complex Between All-D-Enantiomeric Peptide D3 With Wild-Type Amyloid Precursor Protein 672-726 Fragment (Amyloid Beta 1-55)
Organism: Homo sapiens, Synthetic construct
Method: SOLUTION NMR Release Date: 2021-01-13 Classification: PROTEIN BINDING |
 |
Dynamic Complex Between All-D-Enantiomeric Peptide D3 With L723P Mutant Of Amyloid Precursor Protein (App) 672-726 Fragment (Amyloid Beta 1-55)
Organism: Homo sapiens, Synthetic construct
Method: SOLUTION NMR Release Date: 2021-01-13 Classification: PROTEIN BINDING |
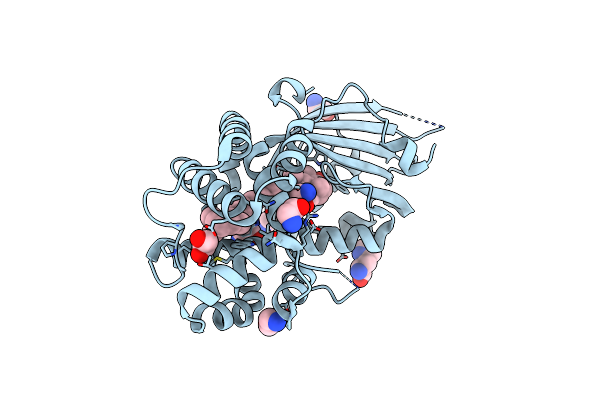 |
Organism: Synechocystis sp. (strain pcc 6803 / kazusa)
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2020-11-18 Classification: PLANT PROTEIN Ligands: ECH, GLY, CL, IMD, HIS, EDO |
 |
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2020-11-18 Classification: PLANT PROTEIN Ligands: ECH, GOL, HIS |
 |
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-11-18 Classification: PLANT PROTEIN Ligands: ASN, ECH, CL, GOL, HIS |
 |
Crystal Structure Of The C14 Ring Of The F1Fo Atp Synthase From Spinach Chloroplast
Organism: Spinacia oleracea
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-12-25 Classification: MEMBRANE PROTEIN Ligands: LFA, OLC |
 |
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2011-05-11 Classification: MEMBRANE PROTEIN Ligands: RET, LI1 |
 |
Structure Of Bacteriorhodopsin Ground State Before And After X-Ray Modification
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2011-05-11 Classification: MEMBRANE PROTEIN Ligands: RET, LI1 |
 |
K Intermediate Structure Of Sensory Rhodopsin Ii/Transducer Complex In Combination With The Ground State Structure
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-03-07 Classification: MEMBRANE PROTEIN Ligands: BOG, RET |
 |
M Intermediate Structure Of Sensory Rhodopsin Ii/Transducer Complex In Combination With The Ground State Structure
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2006-03-07 Classification: MEMBRANE PROTEIN Ligands: BOG, RET |
 |
Molecular Basis Of Transmenbrane Signalling By Sensory Rhodopsin Ii-Transducer Complex
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2002-10-10 Classification: MEMBRANE PROTEIN Ligands: BOG, RET |

