Search Count: 124
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2025-02-12 Classification: RNA BINDING PROTEIN |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2025-02-12 Classification: RNA BINDING PROTEIN Ligands: MN |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2025-02-12 Classification: RNA BINDING PROTEIN |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2025-02-12 Classification: RNA BINDING PROTEIN Ligands: PEG |
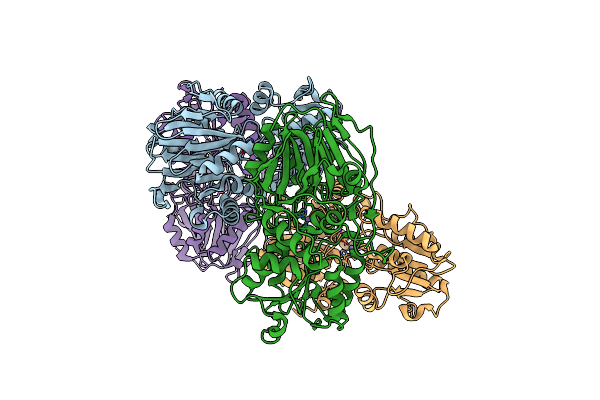 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-02-12 Classification: RNA BINDING PROTEIN Ligands: MN |
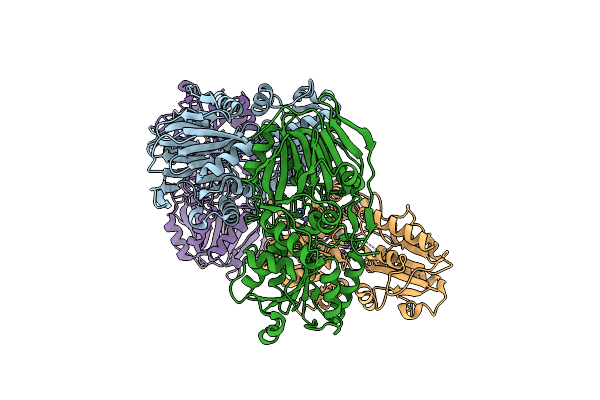 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2025-02-12 Classification: RNA BINDING PROTEIN Ligands: MN |
 |
Ttx183A - A C-Type Cytochrome Domain From The Teredinibacter Turnerae Protein Tertu_2913
Organism: Teredinibacter turnerae
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-03-13 Classification: ELECTRON TRANSPORT Ligands: HEC, CL |
 |
Ttx183B - A C-Type Cytochrome Domain From The Teredinibacter Turnerae Protein Tertu_2913
Organism: Teredinibacter turnerae
Method: X-RAY DIFFRACTION Release Date: 2024-03-13 Classification: ELECTRON TRANSPORT Ligands: HEC, CL, NA |
 |
Se-Met Labelled Ttx122A - A Domain Of Unknown Function From The Teredinibacter Turnerae Protein Tertu_3803
Organism: Teredinibacter turnerae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-03-13 Classification: UNKNOWN FUNCTION Ligands: CA, MG, EDO |
 |
Ttx122A - A Domain Of Unknown Function From The Teredinibacter Turnerae Protein Tertu_3803
Organism: Teredinibacter turnerae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-03-13 Classification: UNKNOWN FUNCTION Ligands: CA, MG, EDO |
 |
Ttx122B - A Domain Of Unknown Function From The Teredinibacter Turnerae Protein Tertu_2913
Organism: Teredinibacter turnerae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-03-13 Classification: UNKNOWN FUNCTION Ligands: CA |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With Barbituric Acid And Citric Acid, Form I
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2023-07-12 Classification: HYDROLASE Ligands: BR8, CIT, EDO, NA |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With Barbituric Acid, Form I
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: BR8, EDO, CL |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With Barbituric Acid, Form Iii
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: BR8, EDO |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With Barbituric Acid, Form Ii
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: BR8, EDO |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With Isoorotic Acid (2,4-Dihydroxypyrimidine-5-Carboxylic Acid), Form I
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: 5CU, EDO |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With Isoorotic Acid (2,4-Dihydroxypyrimidine-5-Carboxylic Acid) And Citric Acid, Form I
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: 5CU, CIT |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With 2,4-Thiazolidinedione, Form I
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: OD3, EDO |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With Uric Acid, Form Iii
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: URC, EDO |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With 5-Fluoroorotic Acid And Citric Acid, Form I
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: FOT, CIT, EDO |

