Search Count: 66
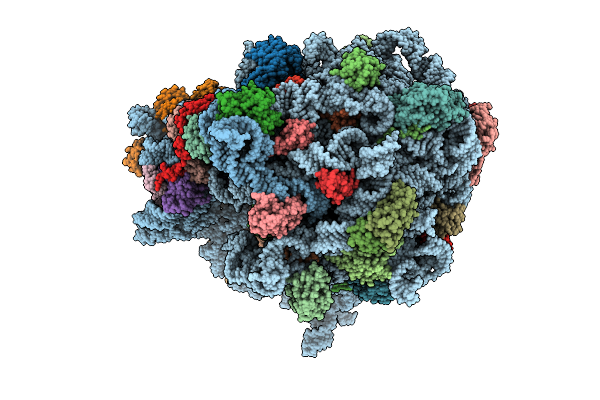 |
E. Coli 70S Ribosome With Unmodified Lys-Trnapro(Ggg) In The P/P Conformation On A Slippery Ccc-C Codon And Elongation Factor P Bound (Ul1 In The Closed Conformation)
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: RIBOSOME Ligands: MG, LYS |
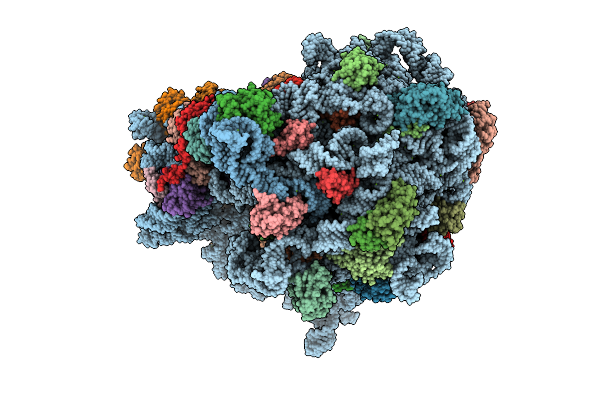 |
E. Coli 70S Ribosome With Unmodified Lys-Trnapro(Ggg) In The P/P Conformation On A Slippery Ccc-C Codon And Elongation Factor P Bound (Ul1 In The Open Conformation)
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: RIBOSOME Ligands: MG |
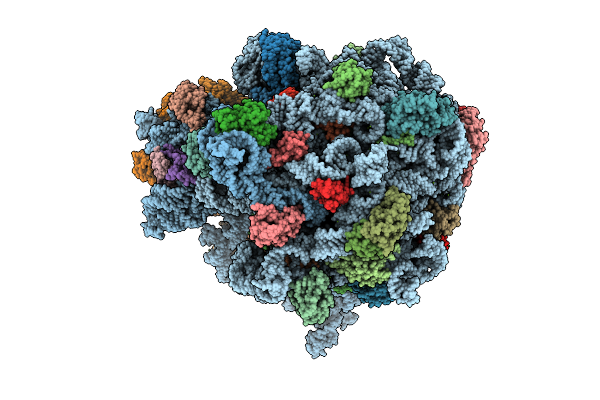 |
E. Coli 70S Ribosome With Unmodified Trnapro(Ggg) In The E*/E Conformation On A Slippery Ccc-C Codon
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: RIBOSOME Ligands: MG |
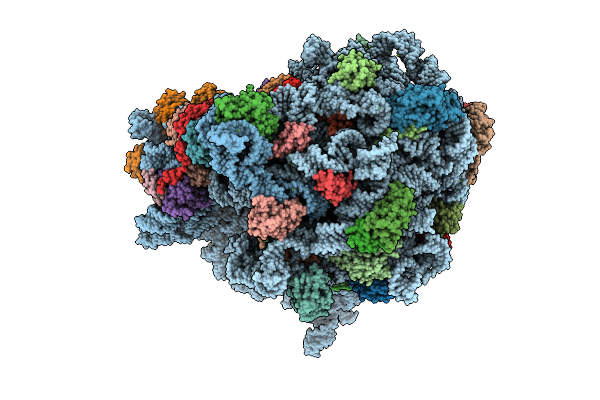 |
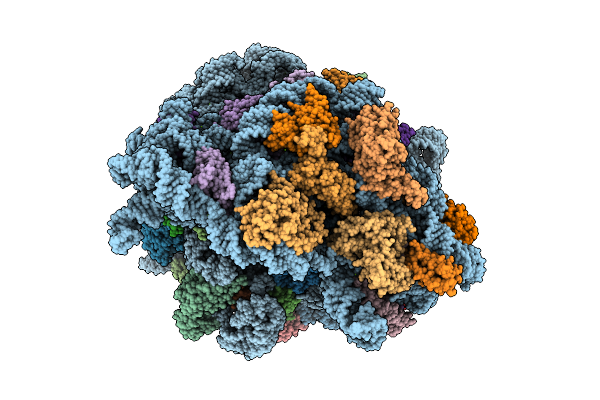
E. Coli 70S Ribosome With Unmodified Lys-Trnapro(Ggg) In The P/P Conformation On A Slippery Ccc-C Codon
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: RIBOSOME Ligands: MG, LYS |
 |
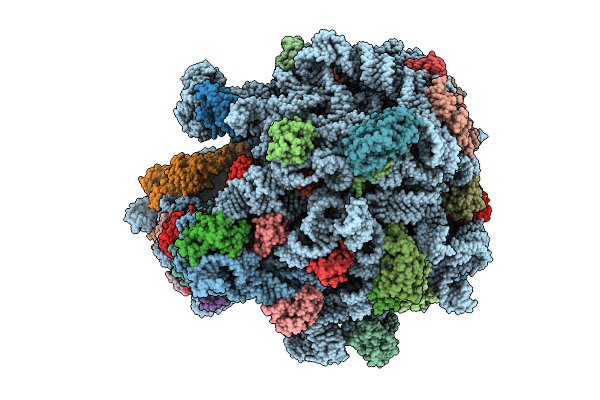
E. Coli 70S Ribosome With Unmodified Trnapro(Ggg) Bound To Slippery P-Site Ccc-C Codon And Trnaval(Uac) In The A Site
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: RIBOSOME Ligands: MG |
 |
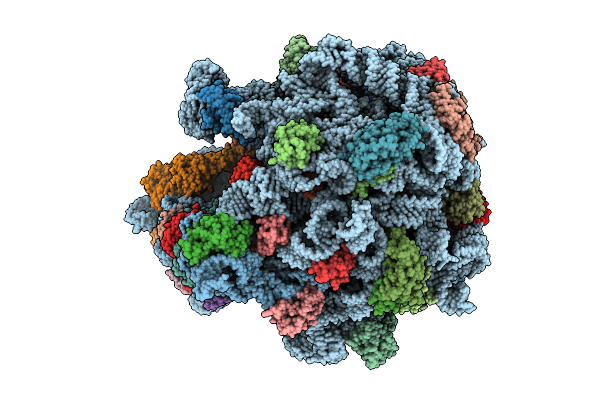
E. Coli 70S Ribosome With Unmodified Lys-Trnapro(Ggg) Bound To Slippery P-Site Ccc-C Codon In The 0 Frame
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: RIBOSOME Ligands: MG, LYS |
 |
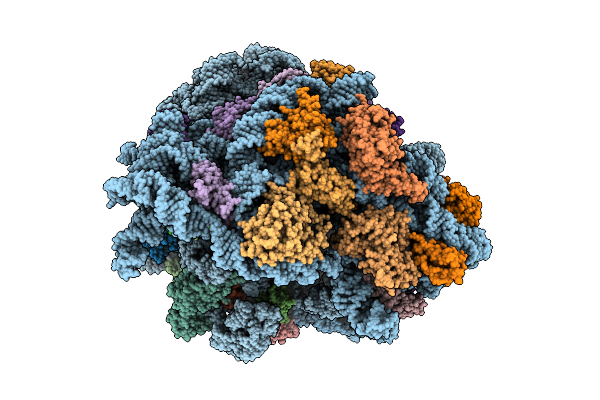
E. Coli 70S Ribosome With Unmodified Lys-Trnapro(Ggg) Bound To Slippery P-Site Ccc-C Codon In The +1 Mrna Reading Frame
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: RIBOSOME Ligands: MG, LYS |
 |
E. Coli 70S Ribosome With Unmodified P/E-Trnapro(Ggg) Bound To Slippery P-Site Ccc-C Codon
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: RIBOSOME Ligands: MG |
 |
E. Coli 70S Ribosome With Unmodified E*/E-Trnapro(Ggg) Bound To Slippery P-Site Ccc-C Codon
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: RIBOSOME Ligands: MG |
 |
E. Coli 70S Ribosome With Unmodified E*/E-Trnapro(Ggg) Bound To Slippery P-Site Ccc-C Codon
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: RIBOSOME Ligands: MG |
 |
E. Coli 70S Ribosome With Unmodified P/E-Trnapro(Ggg) Bound To Slippery P-Site Ccc-C Codon
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: RIBOSOME Ligands: MG |
 |
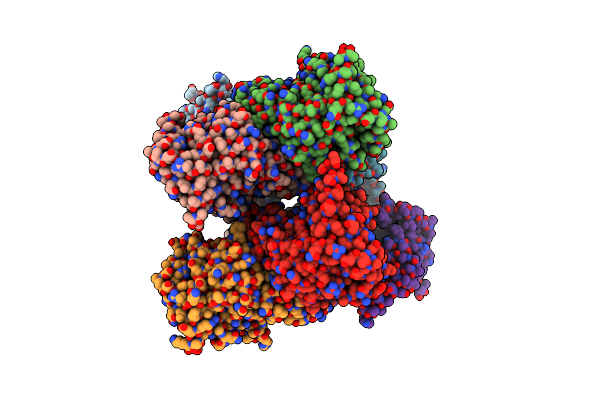
Crystal Structure Of The G11 Protein Heterotrimer Bound To Ym-254890 Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-03-19 Classification: SIGNALING PROTEIN Ligands: GDP, EDO, DAM, HF2, THC, OTH, HL2, MAA, ALA, ACE |
 |
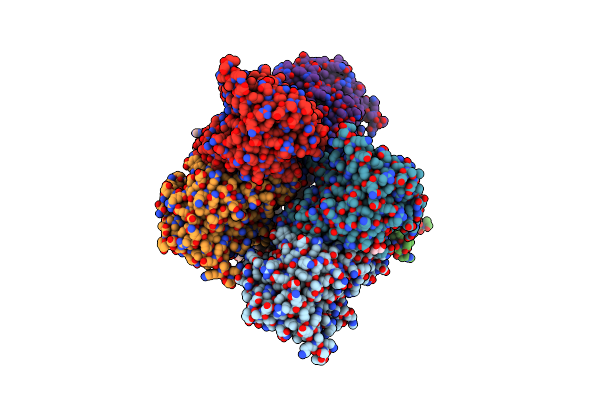
Crystal Structure Of The G11 Protein Heterotrimer Bound To Fr900359 Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2025-03-19 Classification: SIGNALING PROTEIN Ligands: EDO, GDP, ZN, CL, DAM, HF2, UDL, OTH, HL2, MAA, ALA, PPI, DMS |
 |
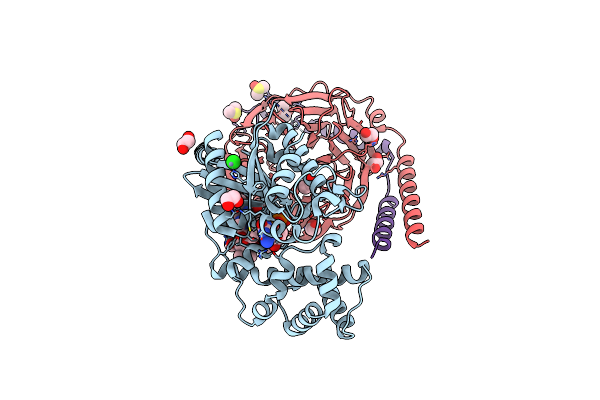
Crystal Structure Of Dimethylsulfone Monooxygenase Sfng From Pseudomonas Fluorescens
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-11-27 Classification: FLAVOPROTEIN Ligands: SO4 |
 |
Crystal Structure Of Dimethylsulfone (Dmso2) Monooxygenase Sfng From Pseudomonas Fluorescens With Dmso2 And Oxidized Fmn Bound
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-11-27 Classification: FLAVOPROTEIN Ligands: FMN, XZ5, GOL, CL |
 |
Crystal Structure Of 2-Hydroxacyl-Coa Lyase/Synthase Apbhacs From Alphaproteobacteria Bacterium In The Complex With Thdp, L-Lactyl-Coa, And Adp
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-10-02 Classification: LYASE Ligands: 8FL, UQ3, ADP, ACE, MG, GOL |
 |
Crystal Structure Of 2-Hydroxacyl-Coa Lyase/Synthase Apbhacs From Alphaproteobacteria Bacterium In The Complex With Thdp, D-Lactyl-Coa, And Adp
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2024-10-02 Classification: LYASE Ligands: MG, 8FL, UQ3, ADP, EDO, ACY, PO4, CL, ACE, PEG |
 |
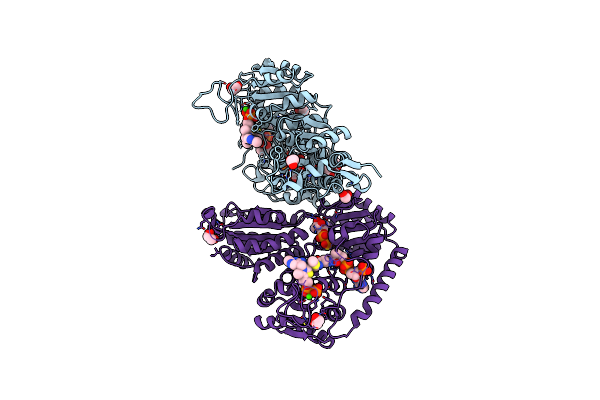
Crystal Structure Of 2-Hydroxyacyl-Coa Lyase/Synthse Apbhacs From Alphaproteobacteria Bacterium In The Complex With Thdp, Formyl-Coa, And Adp
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2024-10-02 Classification: LYASE Ligands: A1AEK, FYN, MG, ADP, EDO, PO4, PEG |
 |
Crystal Structure Of 2-Hydroxyacyl-Coa Lyase/Synthase Tbhacs From Thermoflexaceae Bacterium In The Complex With Thdp, Formyl-Coa And Adp
Organism: Thermoflexaceae bacterium
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-10-02 Classification: LYASE Ligands: TPP, FYN, CA, ADP, EDO, PEG |
 |
Crystal Structure Of 2-Hydroxyacyl-Coa Lyase/Synthase Tbhacs From Thermoflexaceae Bacterium In The Complex With Thdp, 2-Hydroxyisobutyryl-Coa And Adp
Organism: Thermoflexaceae bacterium
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2024-10-02 Classification: LYASE Ligands: A1AEK, COA, ADP, MG, EDO |

