Search Count: 45
 |
Organism: Acinetobacter sp. ver3
Method: X-RAY DIFFRACTION Release Date: 2020-07-22 Classification: OXIDOREDUCTASE Ligands: NAP, HEM, CL, UNX |
 |
Crystal Structure Of V278E-Glyoxalase I Mutant From Zea Mays In Space Group P4(1)2(1)2
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2018-11-21 Classification: PLANT PROTEIN Ligands: GSH, CO, FMT |
 |
Crystal Structure Of V278E-Glyoxalase I Mutant From Zea Mays In Space Group P6(3)
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-11-21 Classification: PLANT PROTEIN Ligands: CO |
 |
Crystal Structure Of E144Q-Glyoxalase I Mutant From Zea Mays In Space Group P4(1)2(1)2
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2018-11-21 Classification: PLANT PROTEIN Ligands: CO, GSH, FMT |
 |
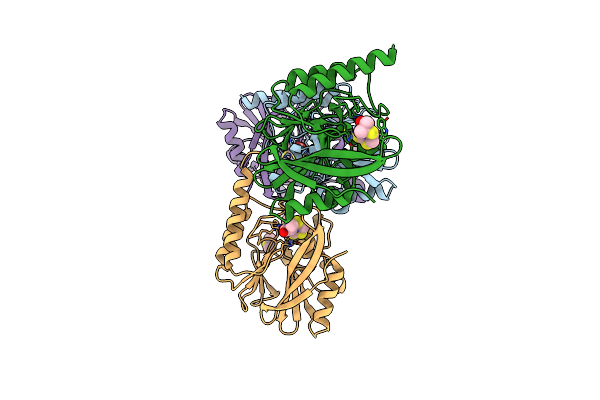
Viral Evolution Results In Multiple, Surface-Allocated Enzymatic Activities In A Fungal Double-Stranded Rna Virus
Organism: Rosellinia necatrix quadrivirus 1
Method: ELECTRON MICROSCOPY Release Date: 2017-11-29 Classification: VIRUS |
 |
Organism: Methylobacterium extorquens (strain atcc 14718 / dsm 1338 / jcm 2805 / ncimb 9133 / am1)
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2017-02-08 Classification: LYASE Ligands: MG, CL, 1PE |
 |
Organism: Methylobacterium extorquens
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2017-02-08 Classification: OXIDOREDUCTASE Ligands: NAD, CA, CL |
 |
Organism: Methylobacterium extorquens
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2017-02-08 Classification: OXIDOREDUCTASE Ligands: CA |
 |
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2016-06-01 Classification: HYDROLASE Ligands: ZN, EDO, ACT |
 |
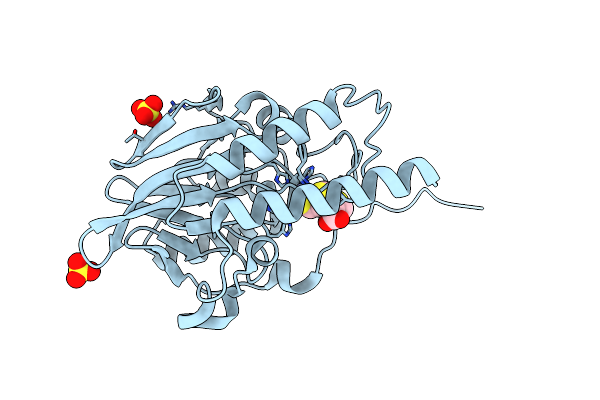
Crystal Structure Of The Metallo-Beta-Lactamase Imp-1 In Complex With The Bisthiazolidine Inhibitor D-Cs319
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-06-01 Classification: HYDROLASE Ligands: ZN, 3R9, EDO, DMS, NA, CL |
 |
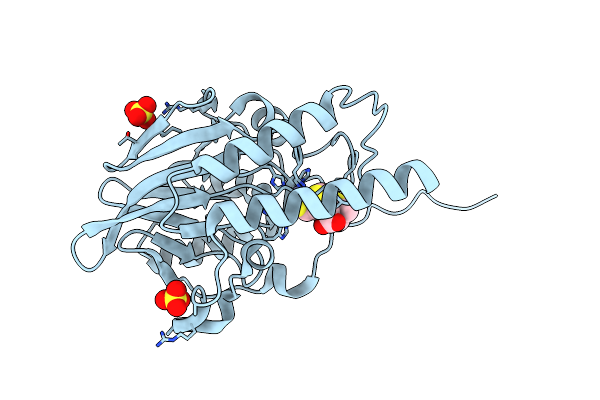
Crystal Structure Of The Metallo-Beta-Lactamase L1 In Complex With The Bisthiazolidine Inhibitor D-Cs319
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2016-06-01 Classification: HYDROLASE Ligands: ZN, 3R9, SO4 |
 |
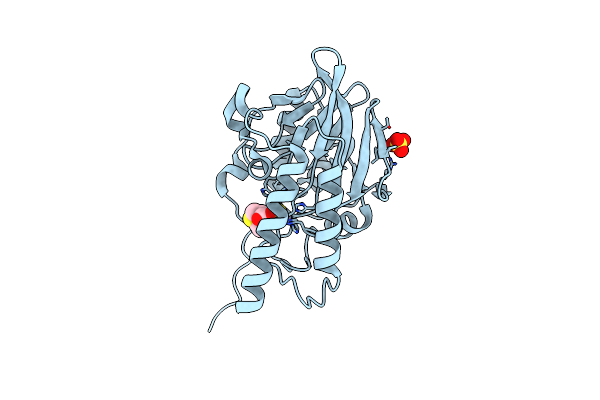
Crystal Structure Of The Metallo-Beta-Lactamase L1 In Complex With The Bisthiazolidine Inhibitor D-Vc26
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-06-01 Classification: HYDROLASE Ligands: VC2, ZN, SO4 |
 |
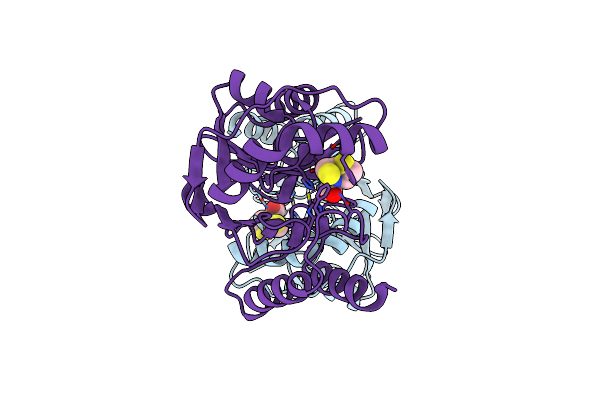
Crystal Structure Of The Metallo-Beta-Lactamase L1 In Complex With The Bisthiazolidine Inhibitor L-Cs319
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2016-06-01 Classification: HYDROLASE Ligands: 3C7, SO4, ZN |
 |
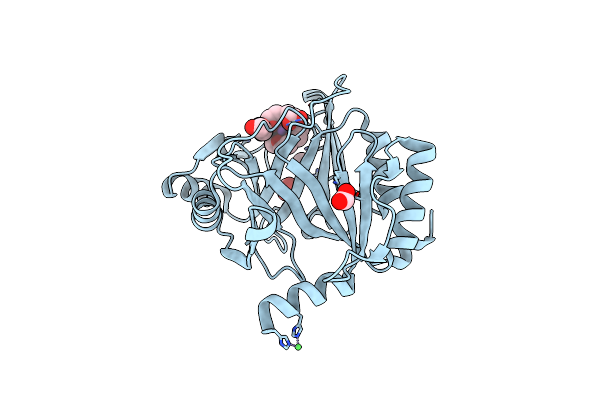
Crystal Structure Of The Metallo-Beta-Lactamase Sfh-I In Complex With The Bisthiazolidine Inhibitor L-Cs319
Organism: Serratia fonticola
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2016-06-01 Classification: HYDROLASE Ligands: ZN, 3C7 |
 |
Crystal Structure Of The Metallo-Beta-Lactamase Imp-1 In Complex With The Bisthiazolidine Inhibitor L-Vc26
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-06-01 Classification: HYDROLASE Ligands: ZN, 9BZ, EDO, DMS |
 |
Crystal Structure Of Enolase From Synechococcus Elongatus, Complex With Phosphoenolpyruvate
Organism: Synechococcus elongatus pcc 7942 = fachb-805
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-04-20 Classification: LYASE Ligands: PEP, CA, ACT |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2015-09-09 Classification: LYASE Ligands: NI, FMT, NA, 2PE |
 |
Organism: Synechococcus elongatus (strain atcc 33912 / pcc 7942 / fachb-805)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2014-12-17 Classification: LYASE Ligands: CA |
 |
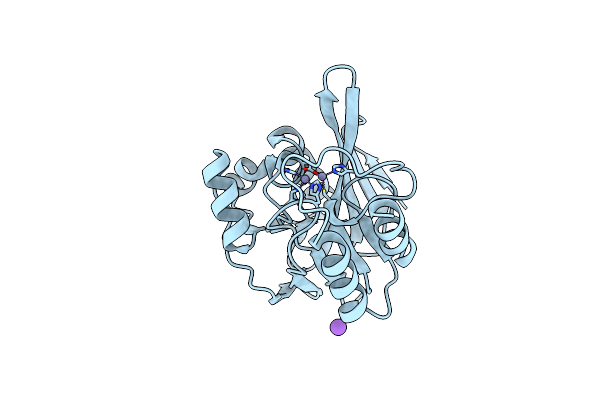
Crystal Structure Of Methylobacterium Extorquens Malate Dehydrogenase Complexed With Oxaloacetate And Adenosine-5-Diphosphoribose
Organism: Methylobacterium extorquens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2014-12-17 Classification: OXIDOREDUCTASE Ligands: CA, OAA, APR |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2014-11-26 Classification: HYDROLASE Ligands: ZN, K |

